7.2 Import / read in data
7.2.1 from CSV
Let’s now import the content of a first file in our environment.
There are several ways we can specify the path / location of a file:
- Using the “absolute path”:
# absolute path
geneexp <- read_csv("/users/sbonnin/DataViz_R_2024/DataViz_source_files-main/files/expression_20genes.csv")- Using the “relative path” (i.e. relative to where the session is currently located), e.g.:
# relative path (this assumes you are in the course folder)
geneexp <- read_csv("DataViz_source_files-main/files/expression_20genes.csv")Because your working directory is /your_home_directory/DataViz_R_2024/, R can find the DataViz_source_files-main without requiring that you specify the whole path (relative vs absolute path).
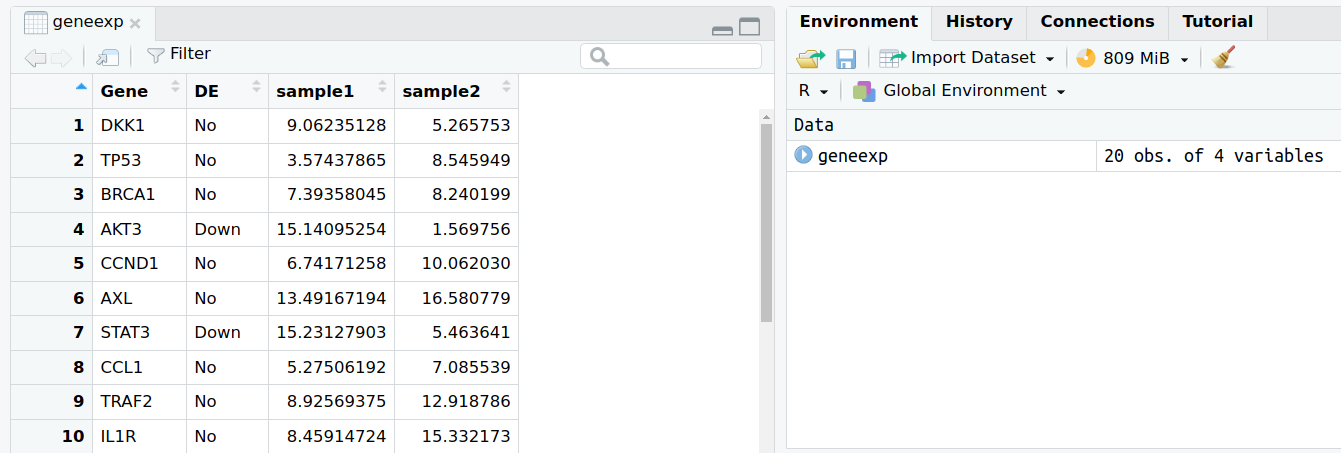
The content of file expression_20genes.csv is now stored in the object named geneexp.
The function also outputs some information about the data you are importing:
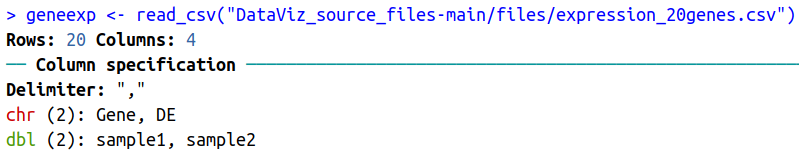
Such as that:
- The data contains 20 rows (observations), and 4 columns (variables).
- Out of these 4 columns:
- 2 contain characters (chr): Gene and DE.
- 2 contain numbers (dbl for “double”): sample1 and sample2
Notes:
- You can find the objects that you create in the Environment tab in the upper-right panel.
- If you click on an object name in the Environment tab, it will open on the upper-left panel. Let’s try with geneexp: