8.12 Faceting
ggplot2 provides facet functions in R, that allow to easily split the plot, according to a given variable.
For example, we can start again from the gtf object.
You can run the following command if you do not have the data loaded in your environment:
We produced a barplot out of this data. However, there is one variable that we did not consider: strand.
Using the faceting function facet_wrap, one can easily split that barplot into 2 plots: one will represent the + strand, one will represent the - strand.
ggplot(data=gtf, mapping=aes(x=chr, fill=gene_type)) +
geom_bar(position="dodge") +
facet_wrap(~strand)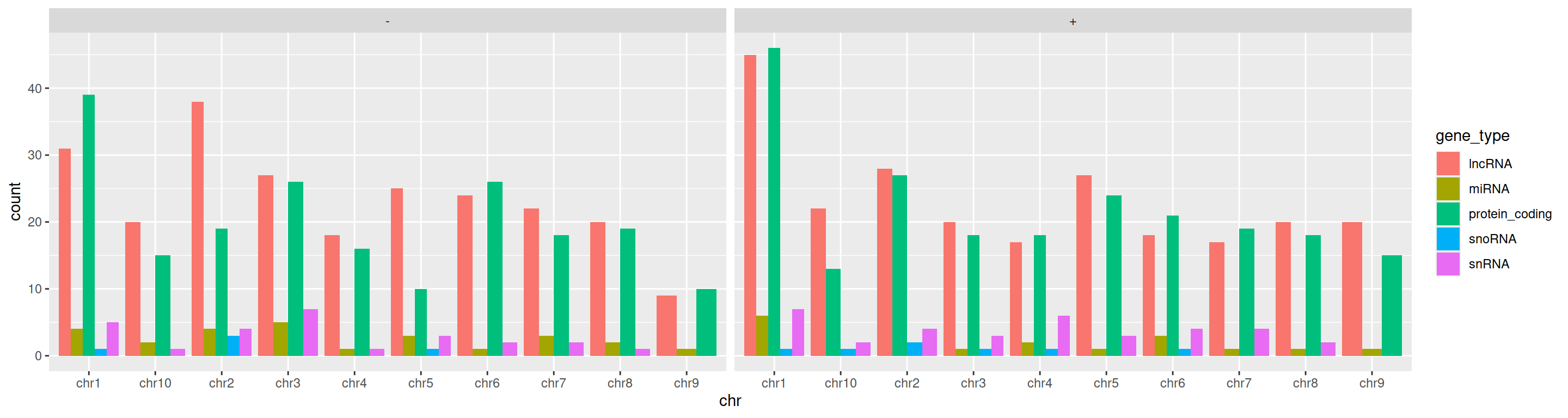
If you want to organize plots vertically, you can set dir=“v”:
ggplot(data=gtf, mapping=aes(x=chr, fill=gene_type)) +
geom_bar(position="dodge") +
facet_wrap(~strand, dir="v")
You can also split/facet the plots using a second variable, for example:
ggplot(data=gtf, mapping=aes(x=chr, color=chr)) +
geom_bar(position="dodge") +
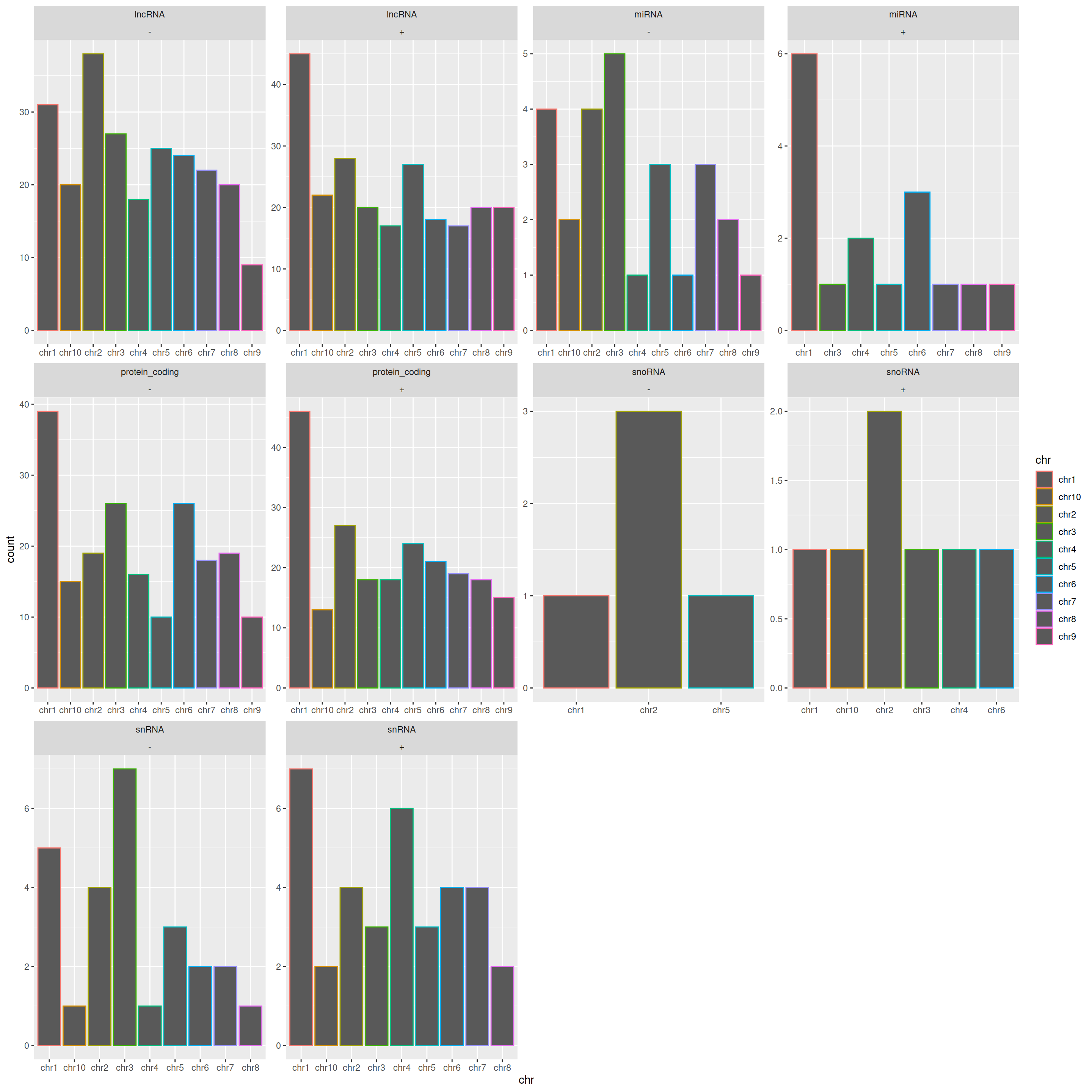
facet_wrap(gene_type~strand)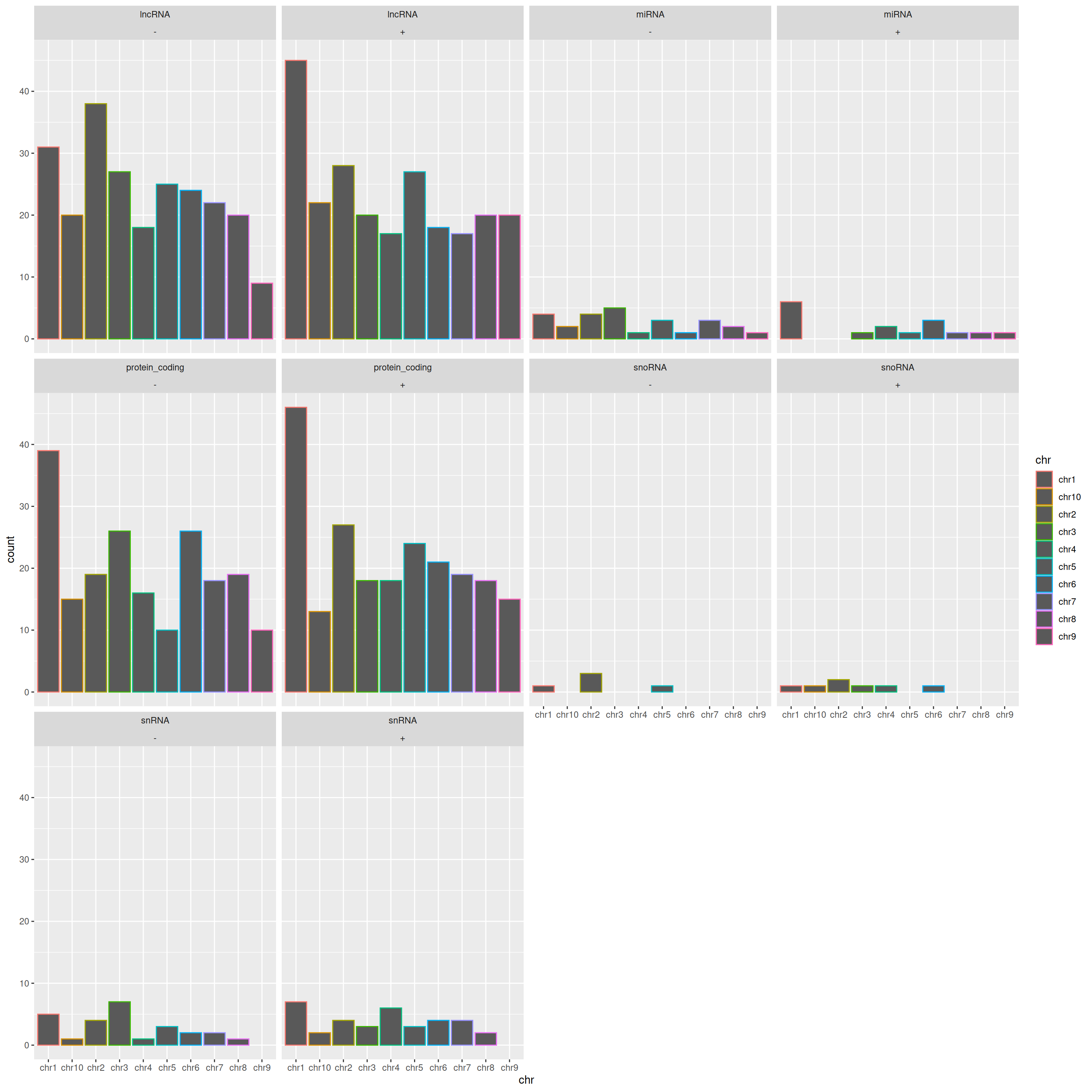
By default, scales are common in all plots. You may want to change this to “free scales” for more readability: it will set the scales per sub-plot.
ggplot(data=gtf, mapping=aes(x=chr, color=chr)) +
geom_bar(position="dodge") +
facet_wrap(gene_type~strand, scales = "free")