10.1 Histograms and density plots
10.1.1 Histogram
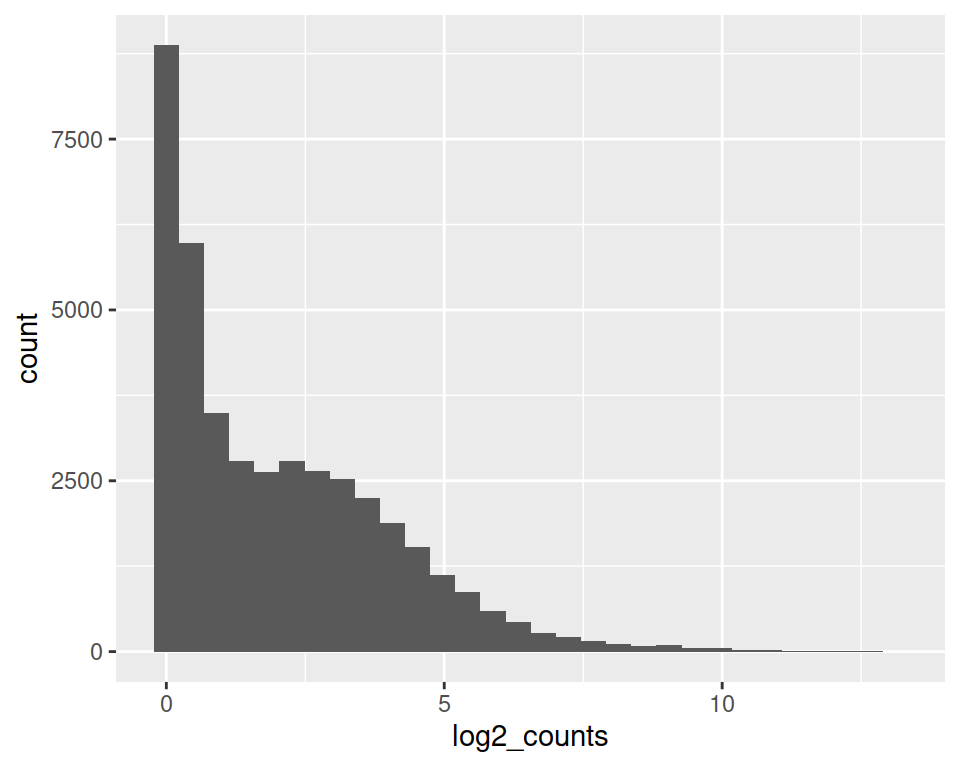
We can use one of our previous objects to create a histogram. You can get it back with:
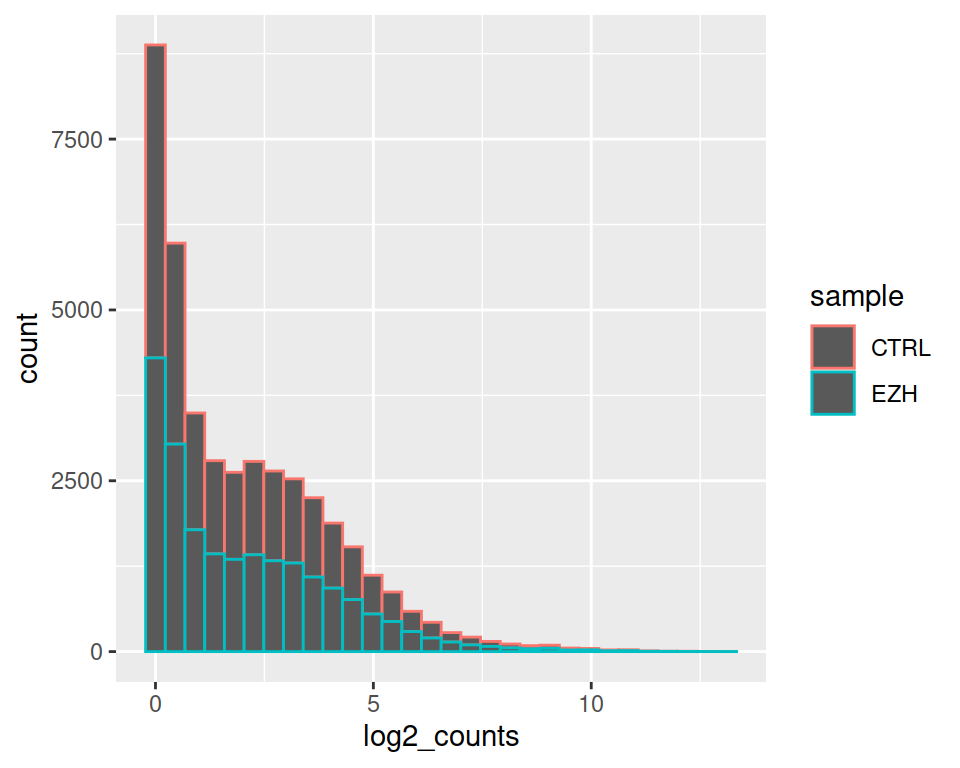
We can split the histogram per sample:
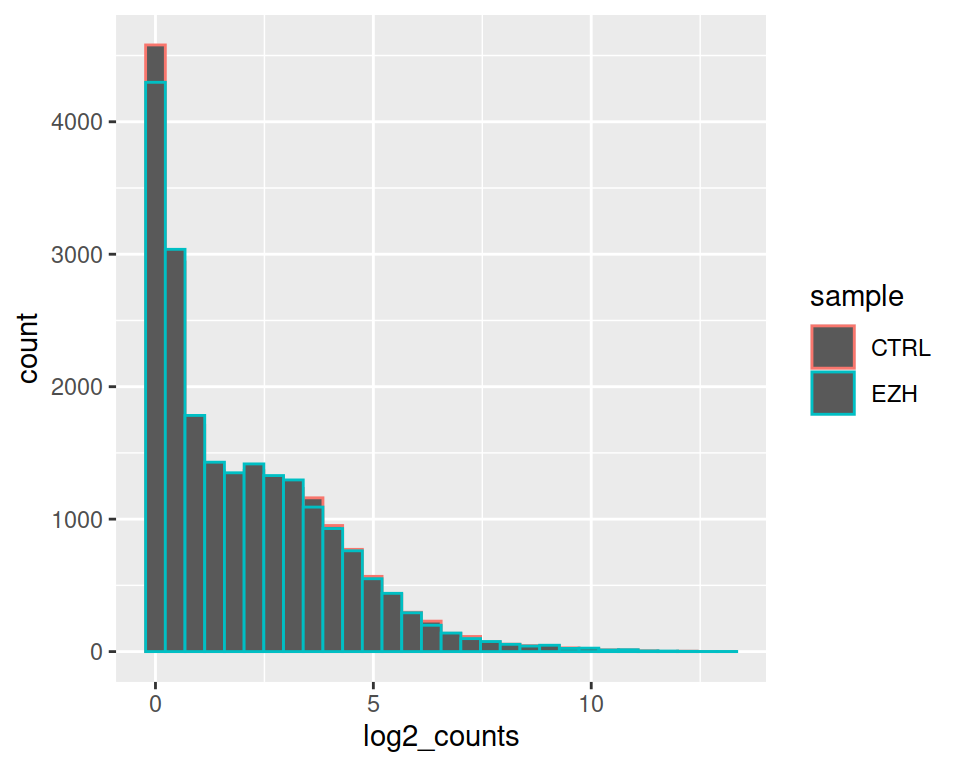
Set position to “identity”, so histograms will not be on top of each other:
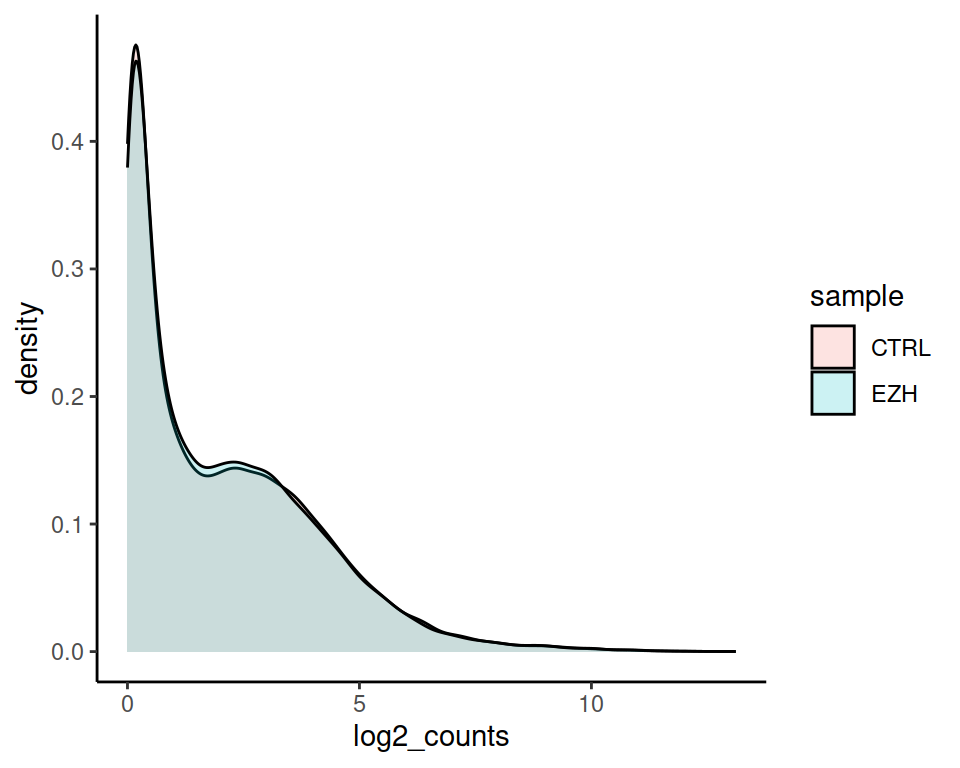
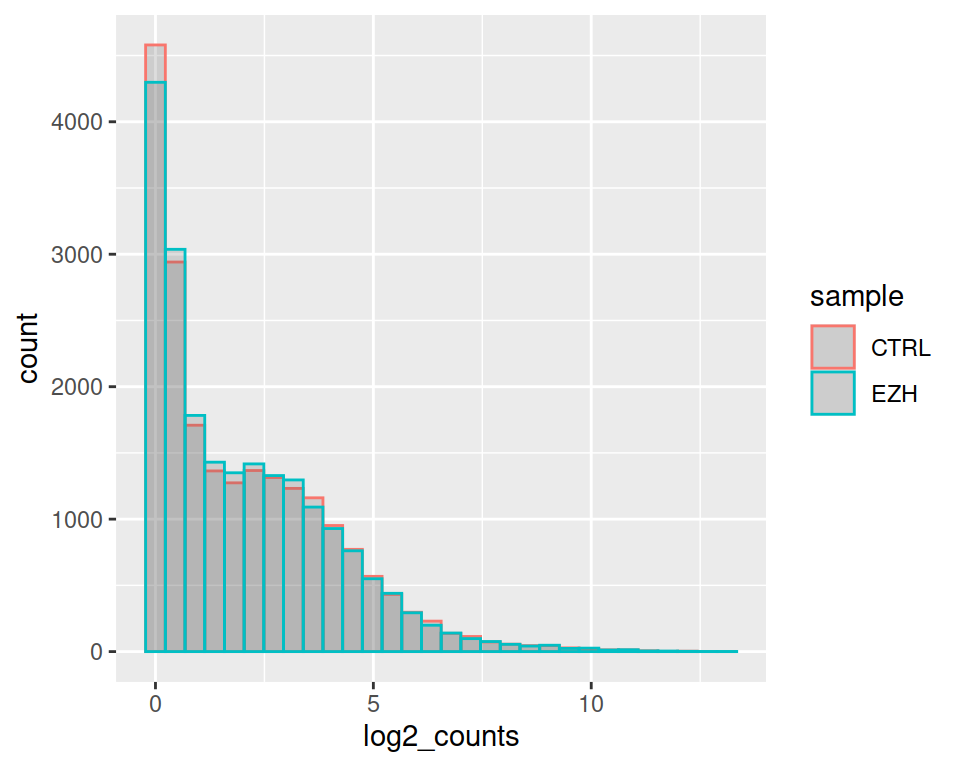
Set alpha (transparency to 0.2):
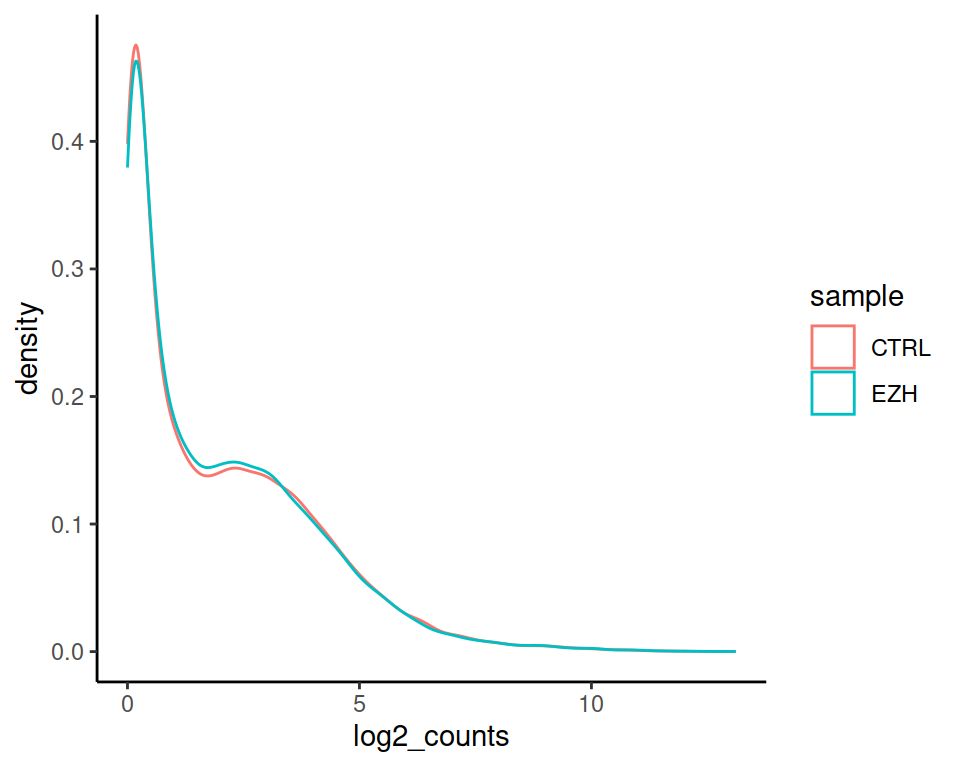
Express as density instead of counts:
ggplot(rnaseq2, aes(x=log2_counts, color=sample)) +
geom_histogram(aes(y=after_stat(density)), position="identity", alpha=0.2) +
theme_classic()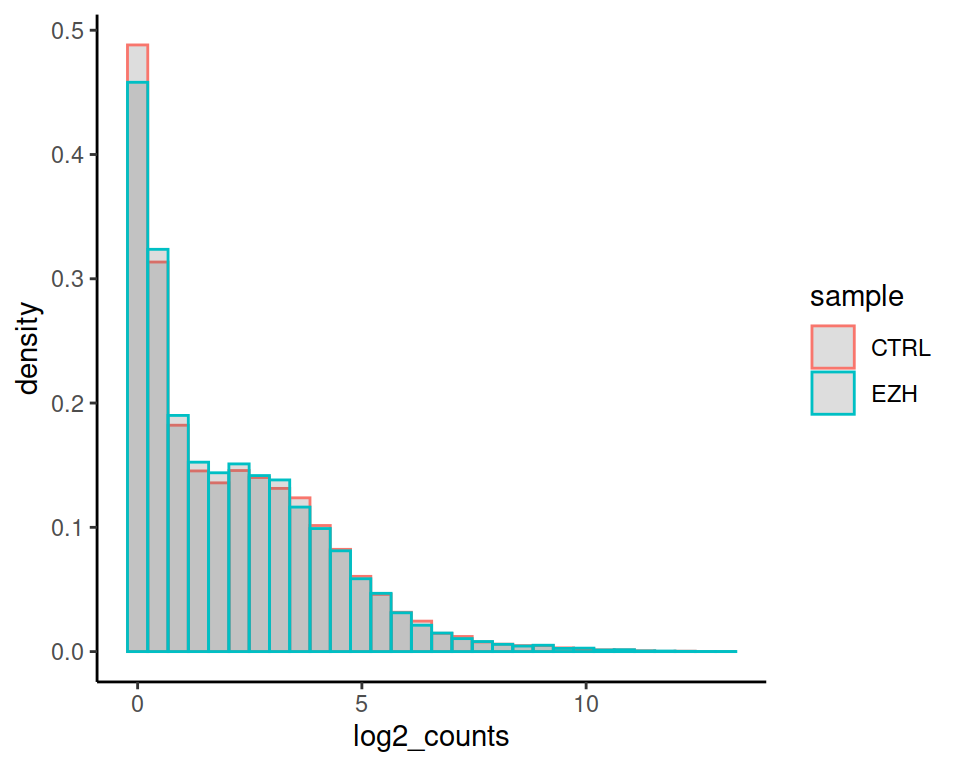
10.1.3 Histogram + density
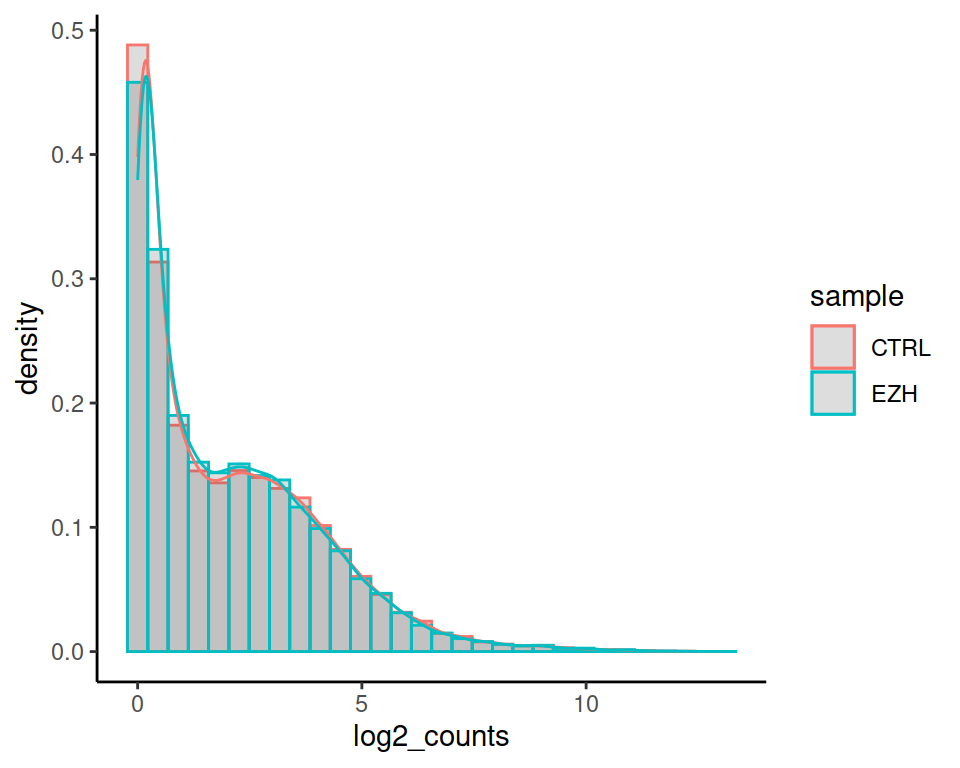
As we combined geom_boxplot and geom_violin in a previous exercise, we can also combine geom_histogram and geom_density:
ggplot(rnaseq2, aes(x=log2_counts, color=sample)) +
geom_histogram(aes(y=after_stat(density)), position="identity", alpha=0.2) +
geom_density(alpha=0.2) +
theme_classic()