10.2 Pie chart
We will plot a pie chart representing the number of genes from gtf found in each chromosome.
Prepare data so as to obtain counts (that is more advanced dplyr manipulation):
The coord_polar() is needed here: pie plots are stacked barplots in polar coordinates.
ggplot(data=gtf_count_chr, mapping=aes(x="", y=counts, fill=chr)) +
geom_bar(stat="identity") +
coord_polar("y", start=0)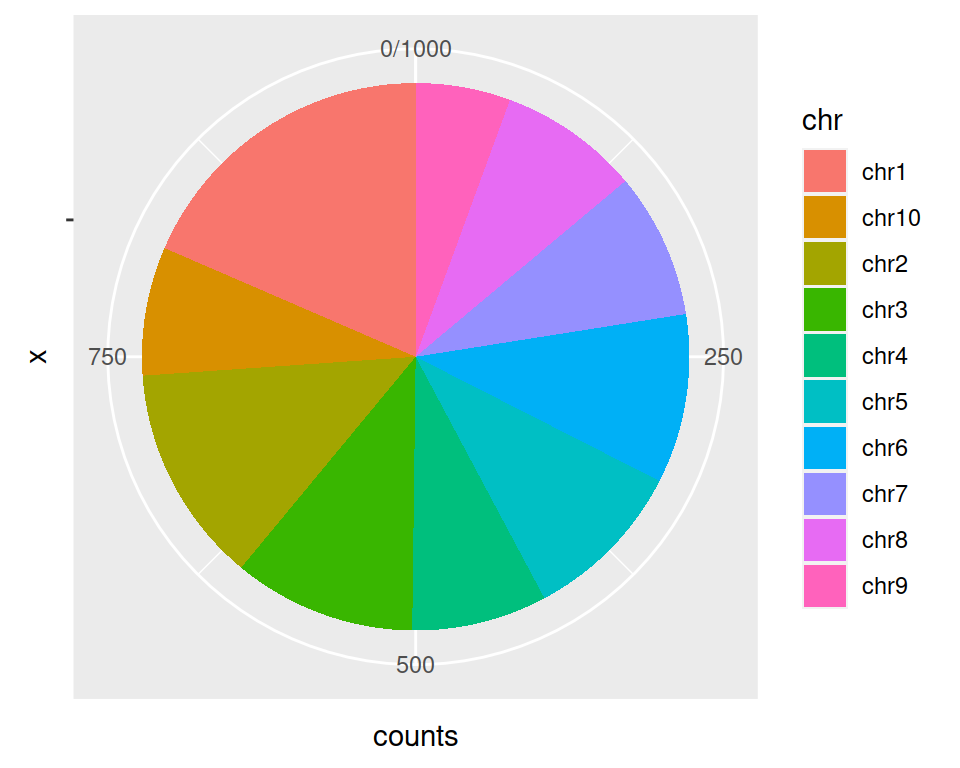
ggplot(data=gtf_count_chr, mapping=aes(x="", y=counts, fill=chr)) +
geom_bar(stat="identity") +
coord_polar("y", start=0) +
theme_void()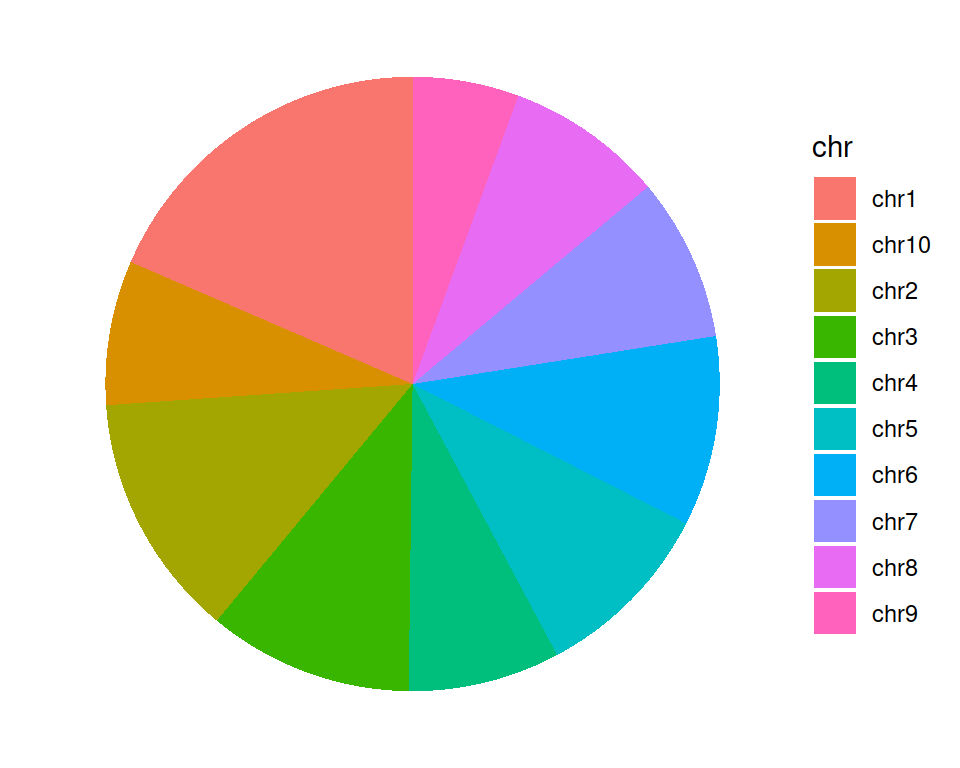
Change the color scale:
ggplot(data=gtf_count_chr, mapping=aes(x="", y=counts, fill=chr)) +
geom_bar(stat="identity") +
coord_polar("y", start=0) +
scale_fill_brewer(palette="Paired") +
theme_void()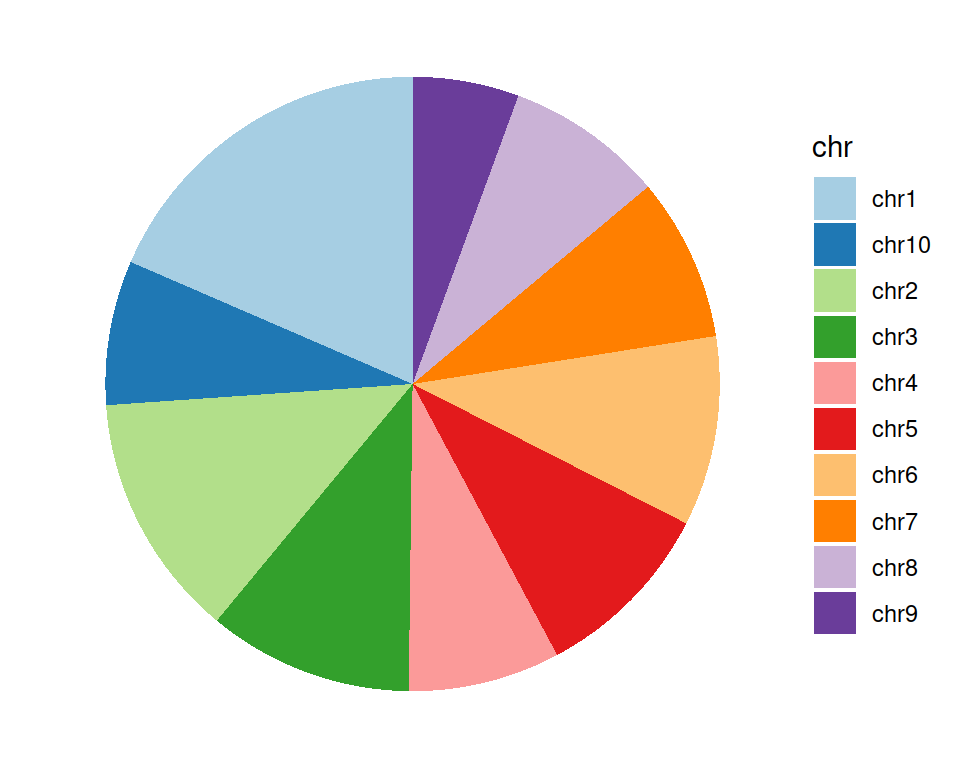
Here is a nice page about more pie plot customization.