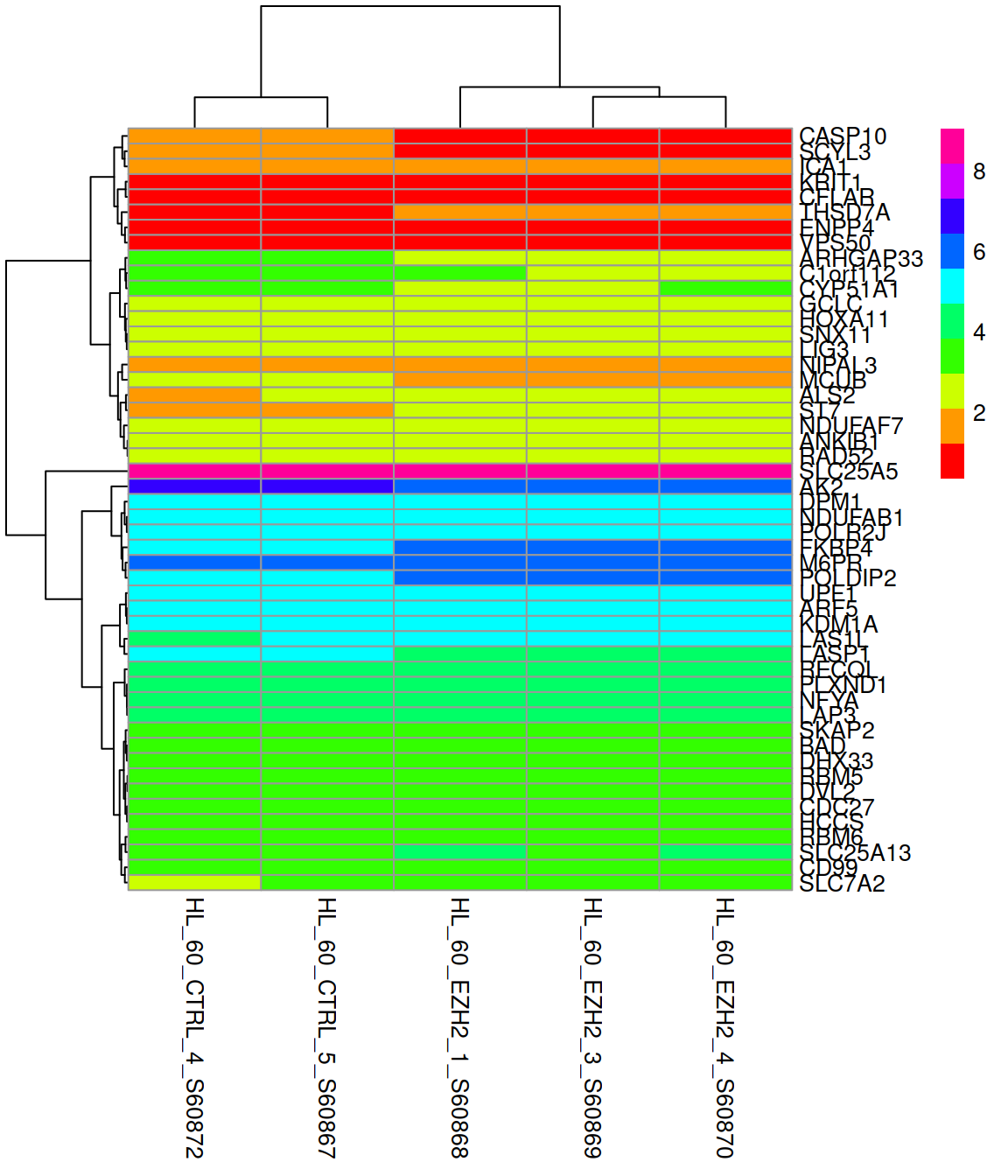
Part13 Heatmaps with {pheatmap}
We will introduce package {pheatmap} to produce heatmaps.
First we can install pheatmap (bottom-right panel -> Packages -> Install).
And then we load the package: tick the package in the list, or load it from the console:
pheatmap is not from the tidyverse package, and does not follow the same structure. It does not work with layers, but with multiple parameters inside a same function.
Let’s read the following data file, and convert it to matrix (that is the input format for pheatmap):
expr_heatmap <- read_csv("DataViz_source_files-main/files/GSE150029_rnaseq_for_heatmap.csv") %>%
column_to_rownames(var = "gene_name") %>%
as.matrix()You can then simply plot the heatmap, giving the matrix as an input:
Control what is clusters: rows, columns, both, or none:
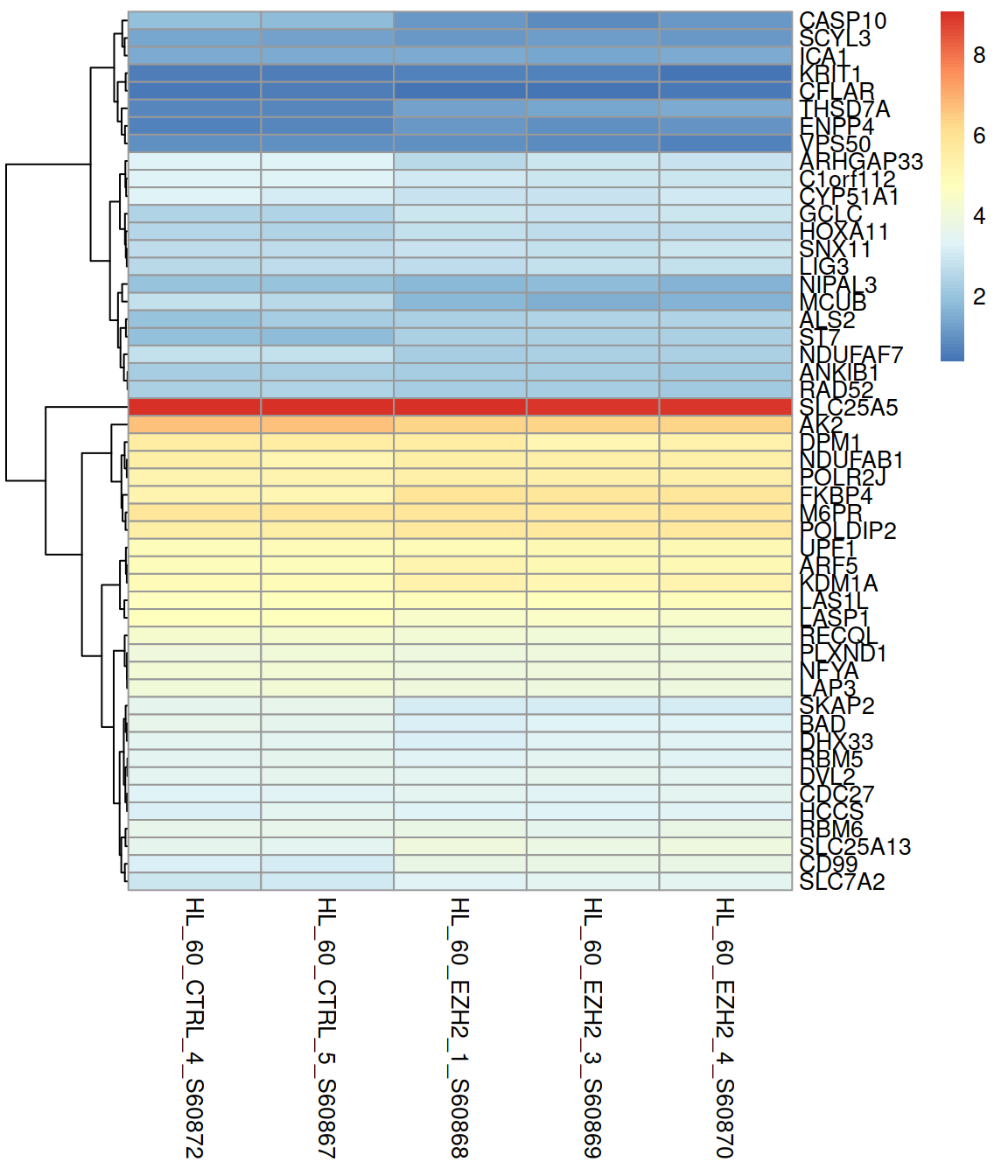
Show or hide row names:
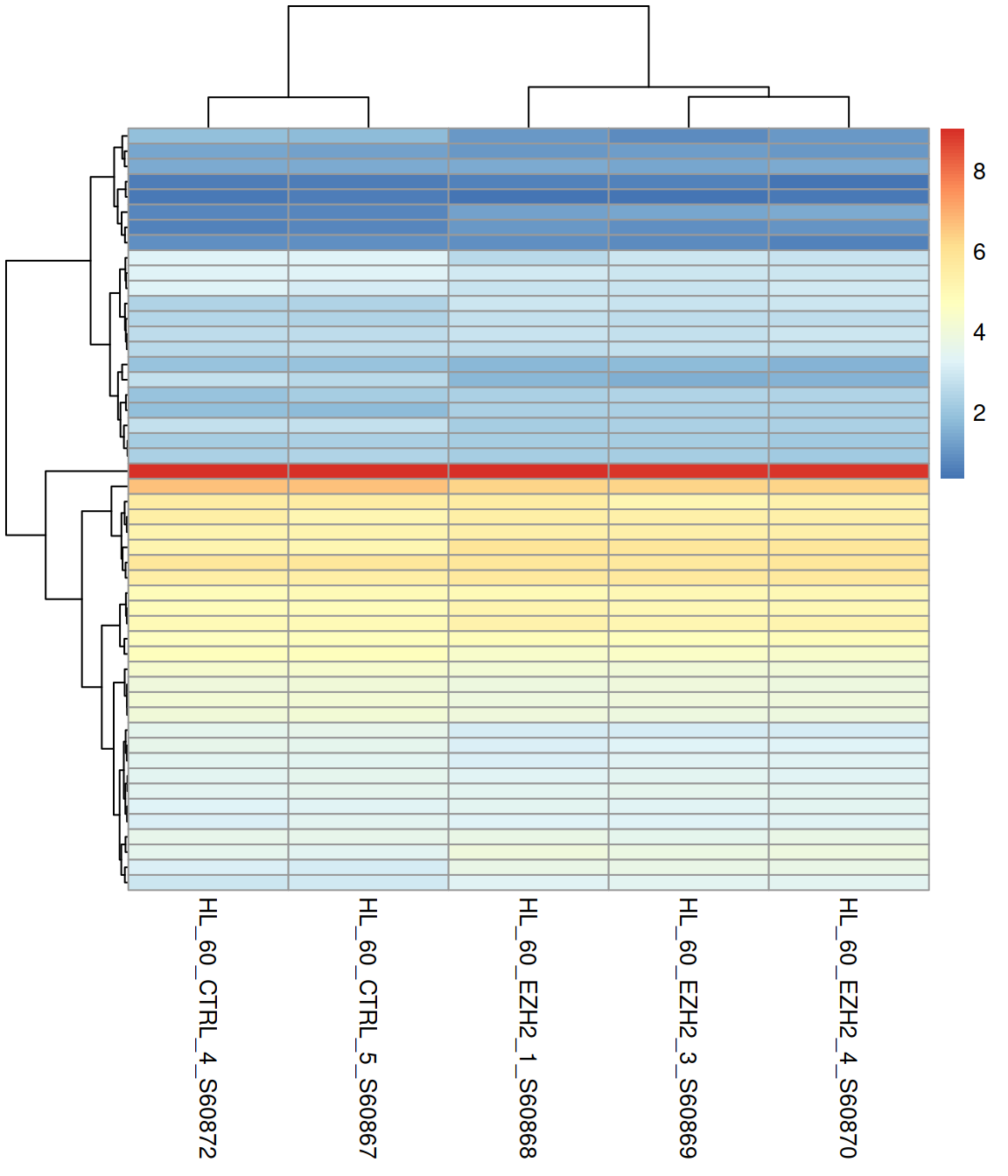
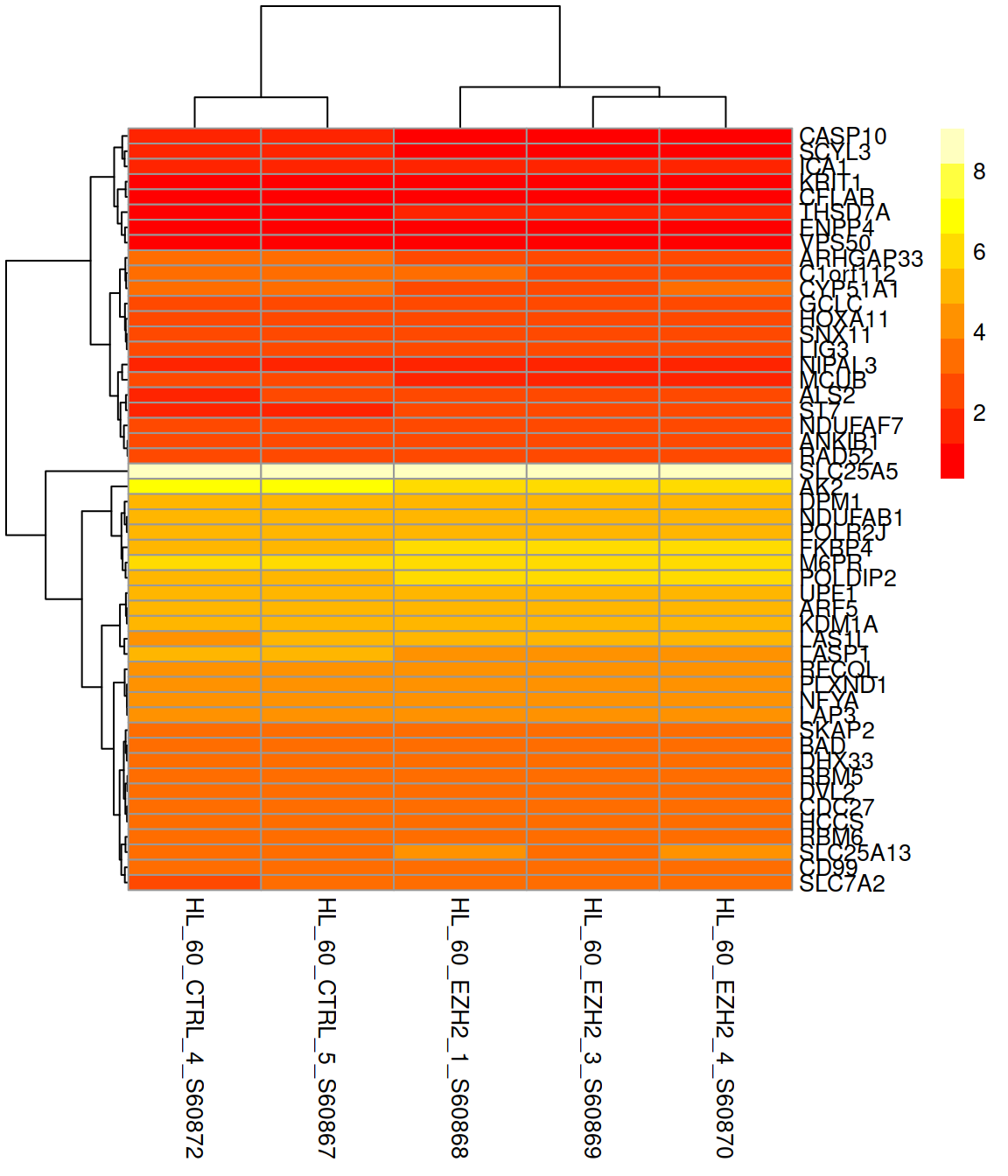
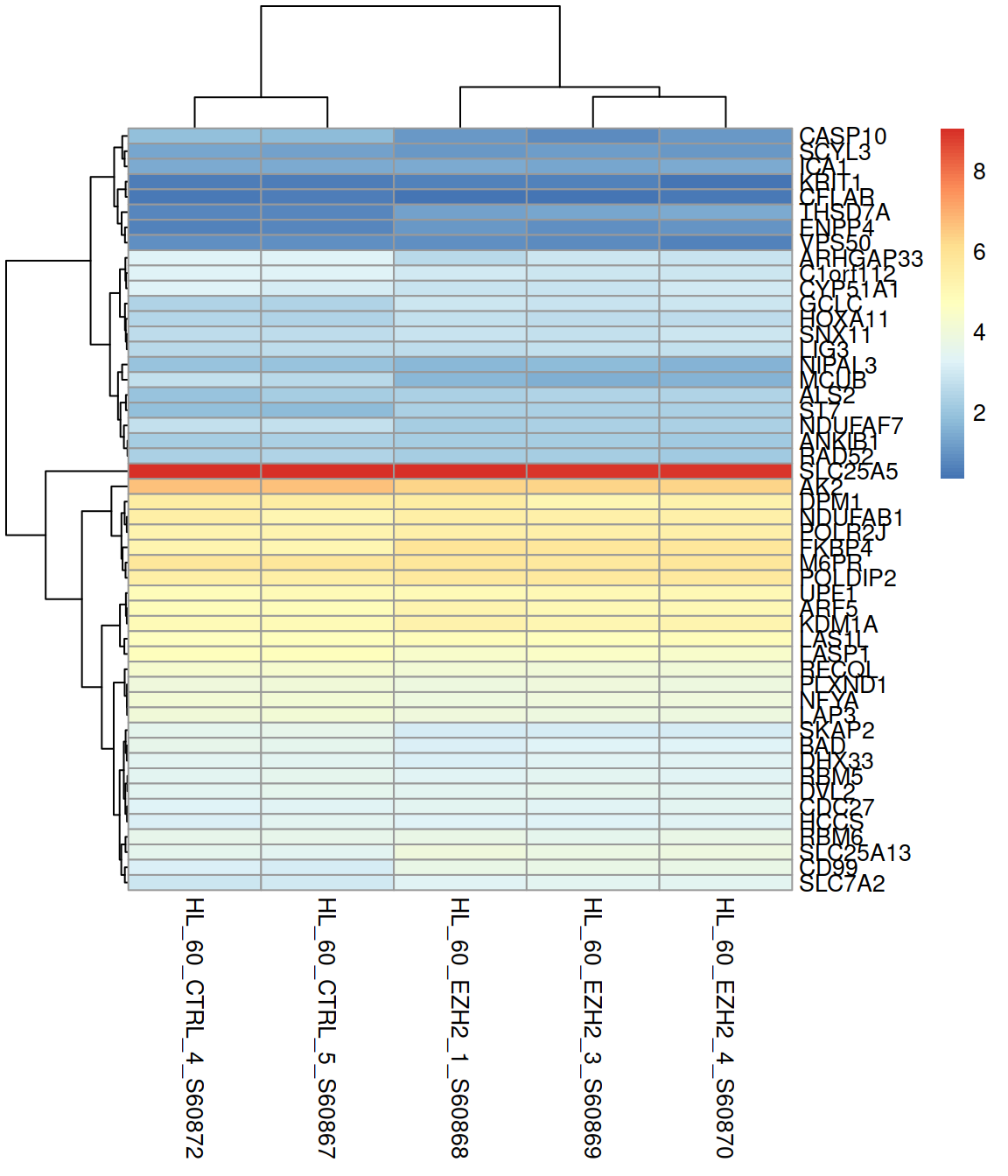
Change the color scale (rainbow() and heat.colors() are available from the base packages):