17.3 Exercise 11: Base plots
Create the script “exercise11.R” and save it to the “Rcourse/Module3” directory: you will save all the commands of exercise 11 in that script.
Remember you can comment the code using #.
correction
17.3.1 Exercise 11a- scatter plot
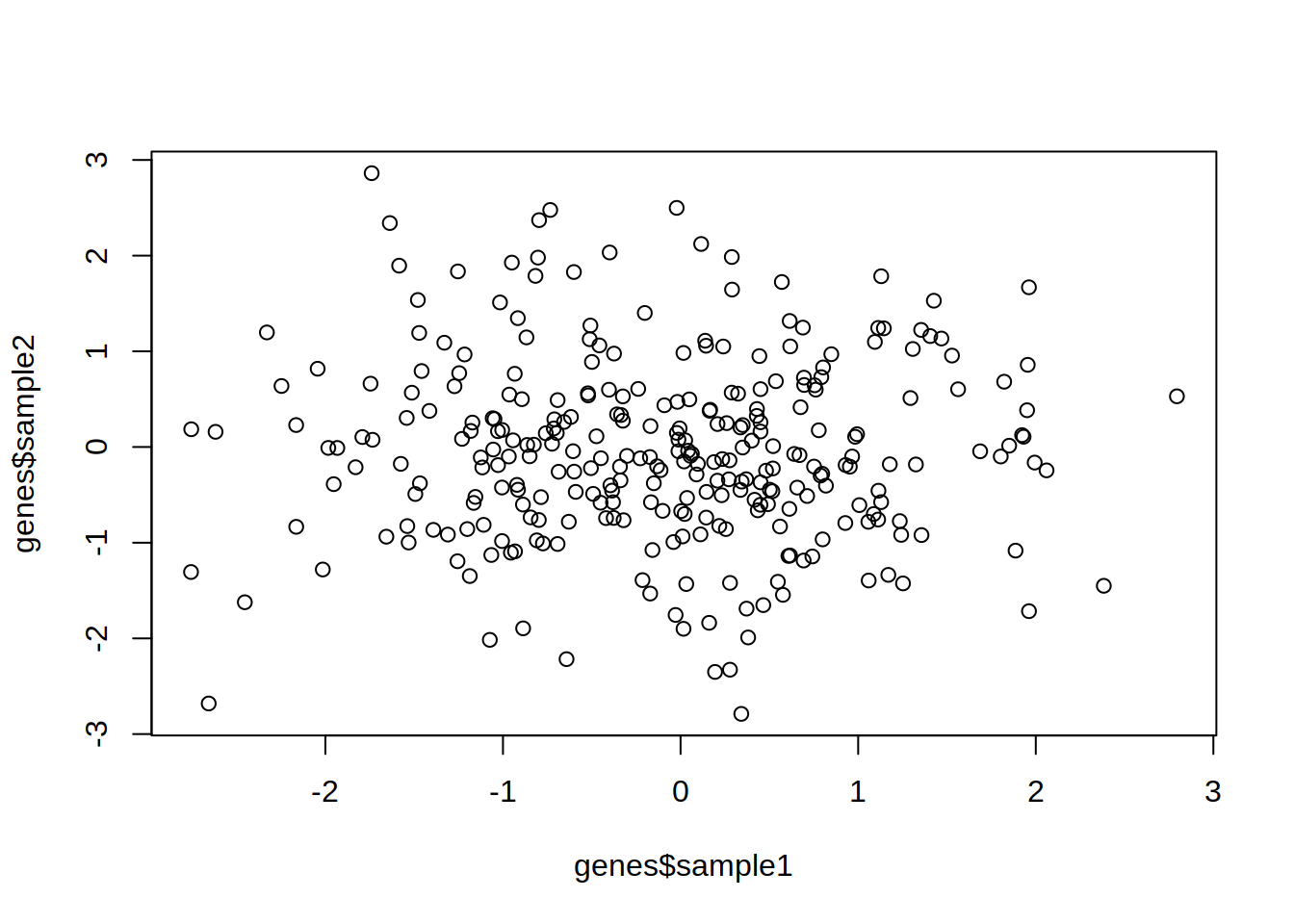
1- Create the following data frame
2- Create a scatter plot showing sample1 (x-axis) vs sample2 (y-axis) of genes.
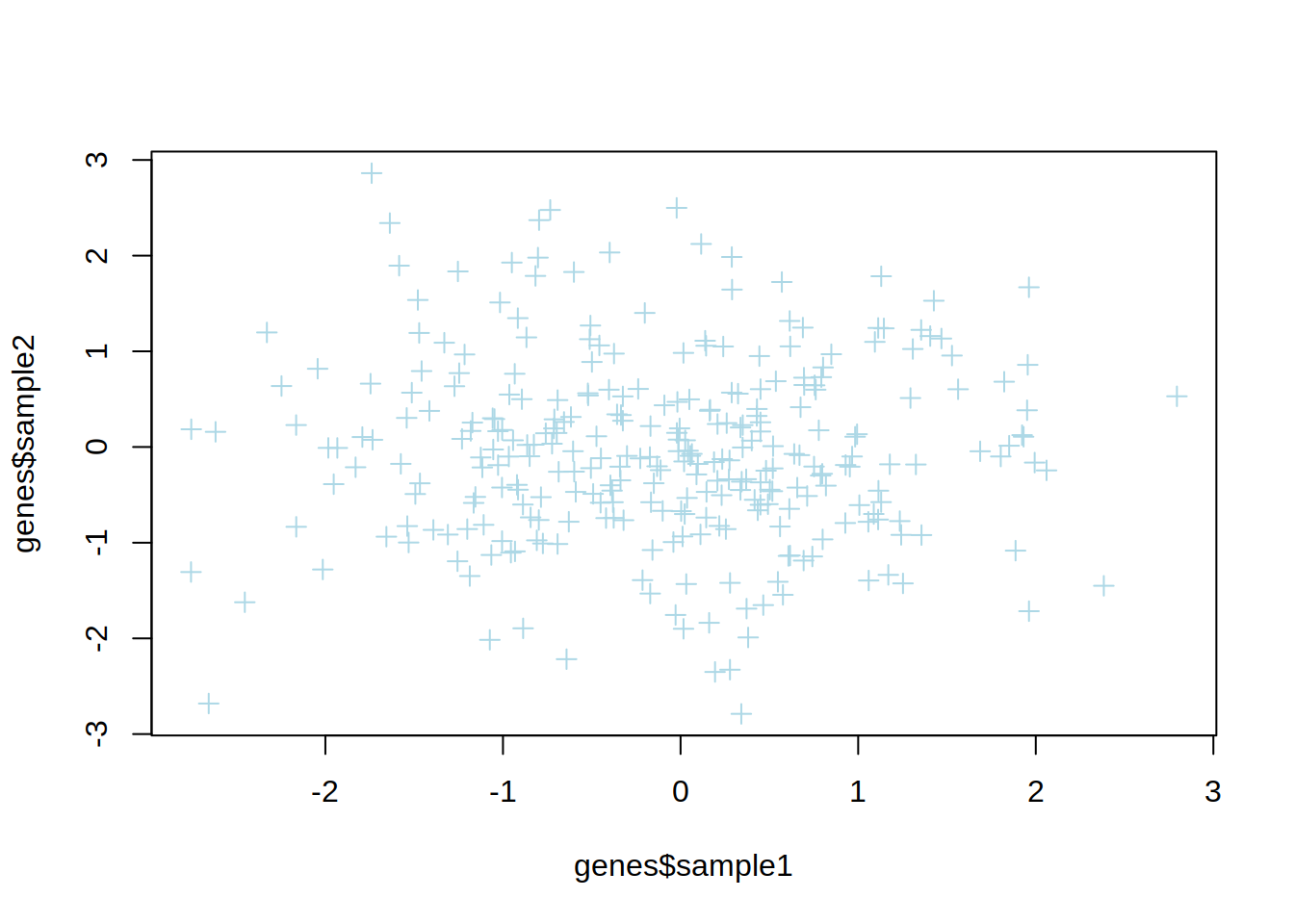
3- Change the point type and color.
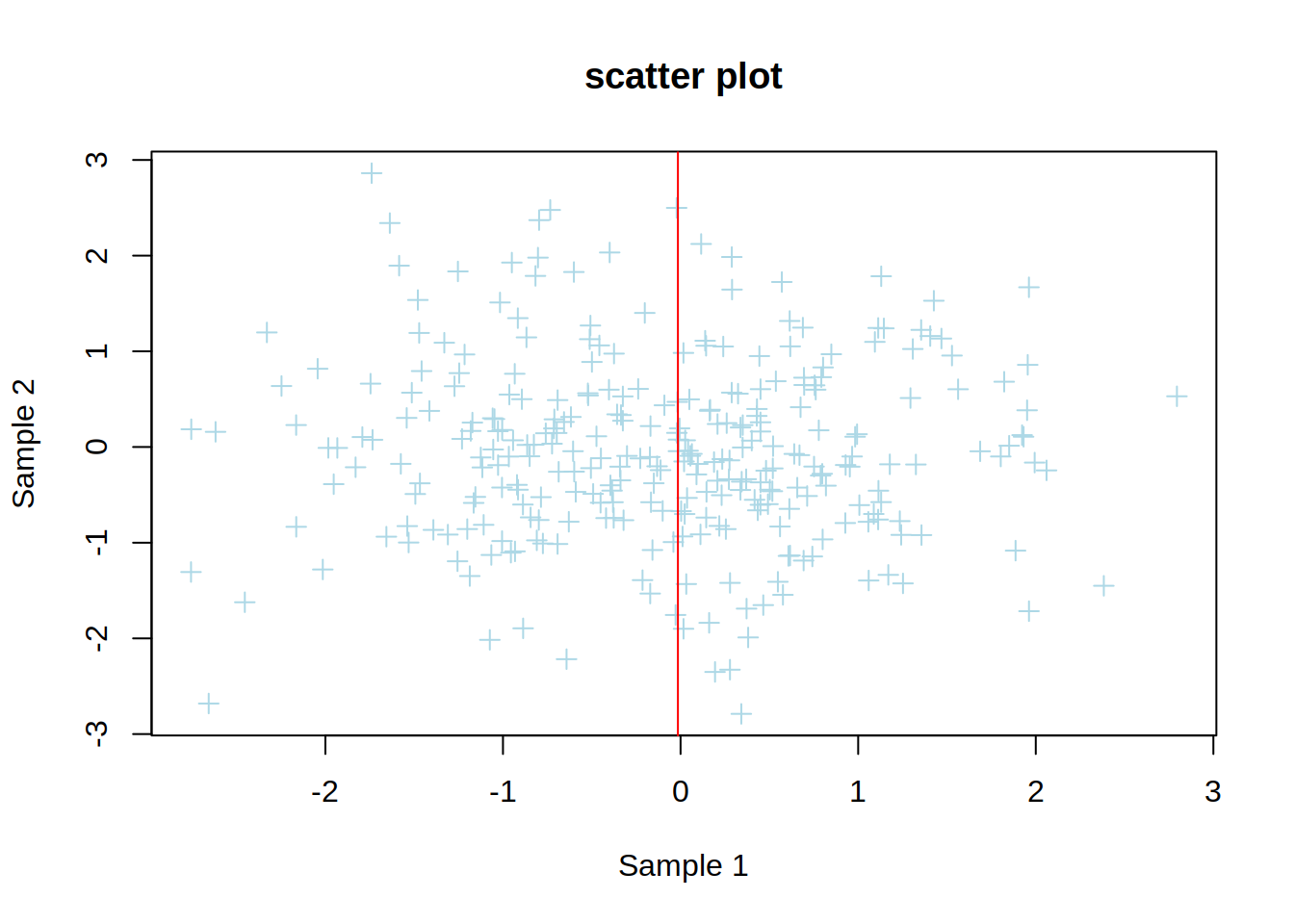
4- Change x-axis and y-axis labels to “Sample 1” and “Sample 2”, respectively.
correction
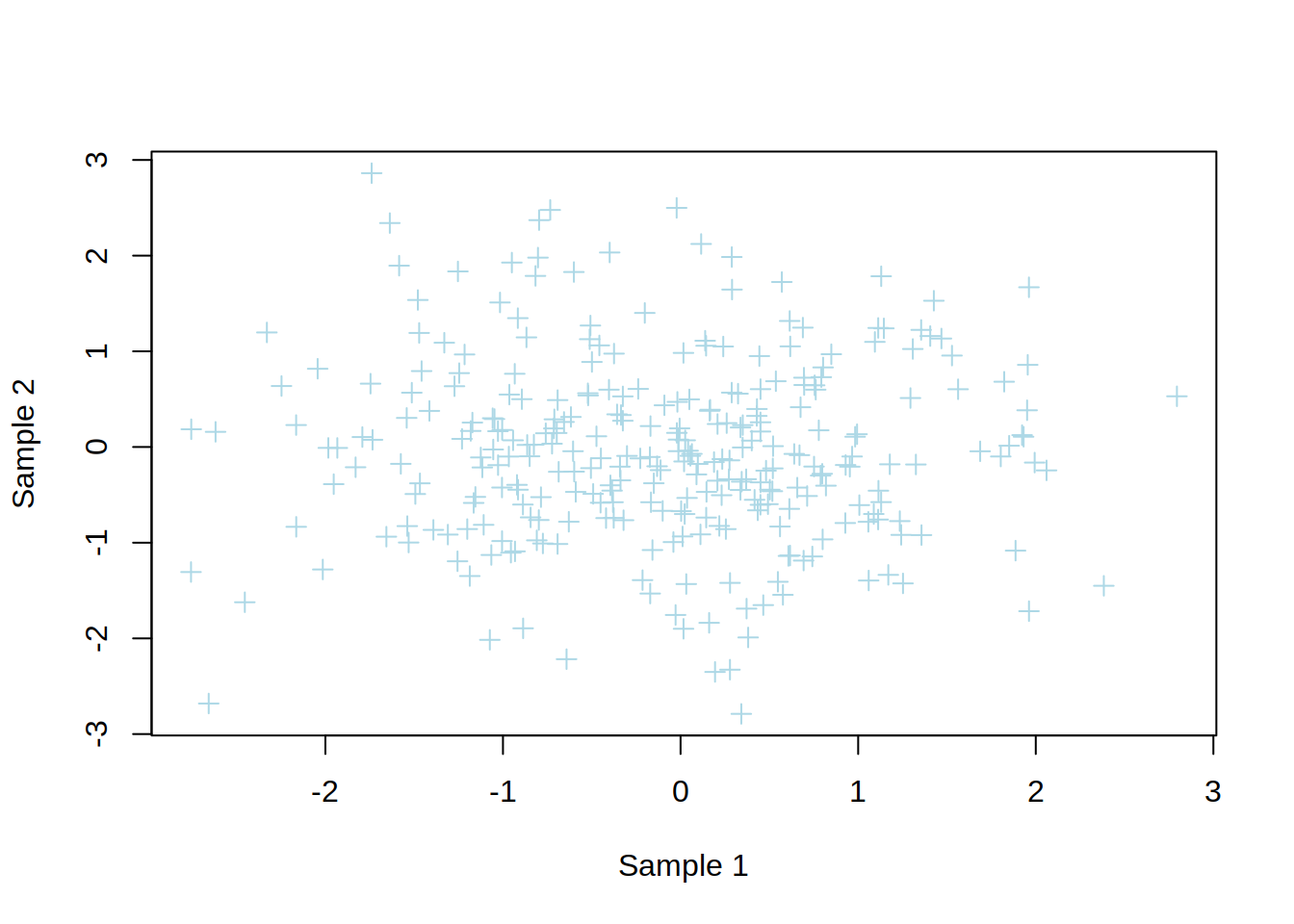
5- Add a title to the plot.
correction
plot(genes$sample1,
genes$sample2,
col="lightblue",
pch=3,
xlab="Sample 1",
ylab="Sample 2",
main="scatter plot")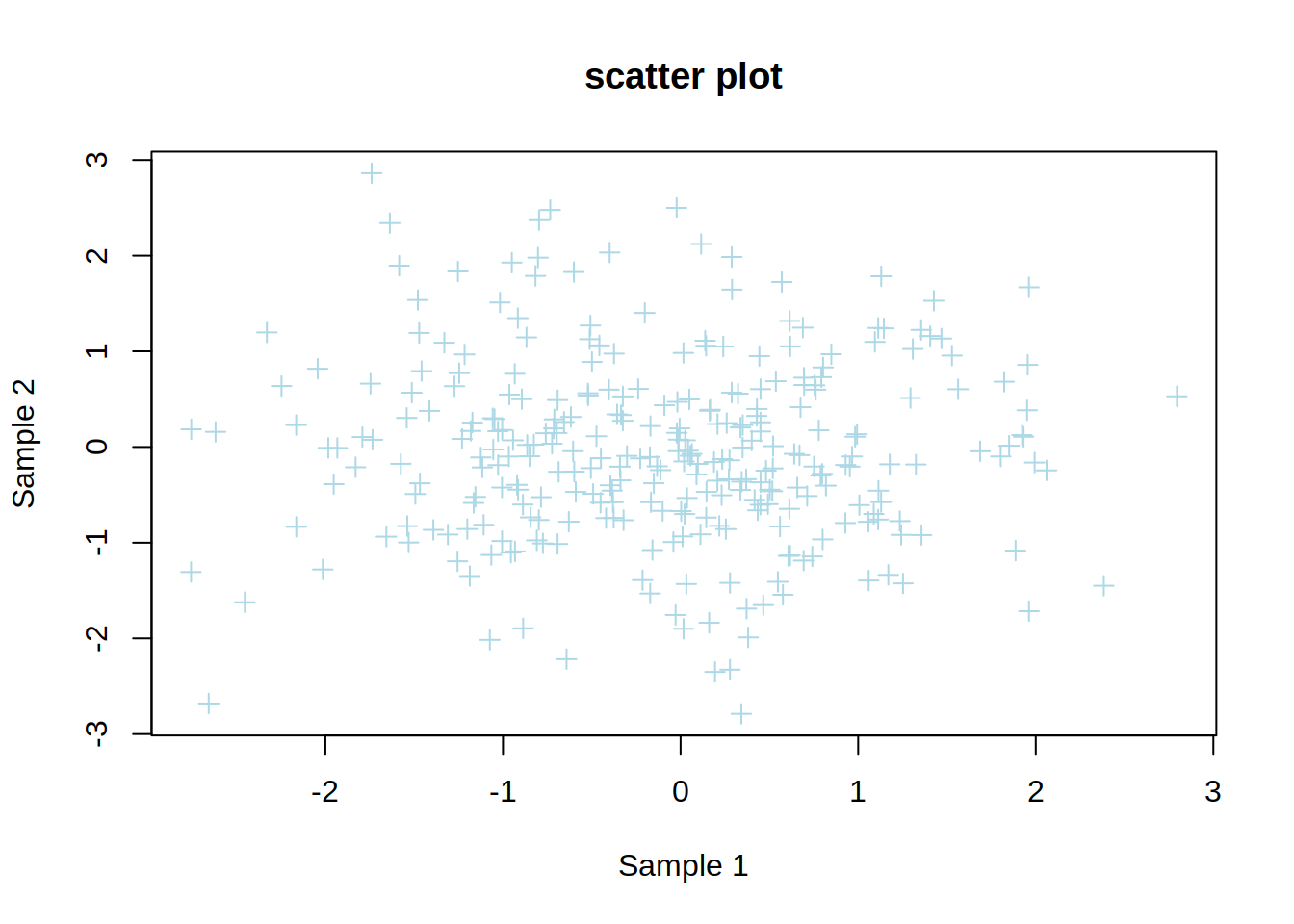
6- Add a vertical red line that starts at the median expression value of sample 1. Do it in two steps:
a. calculate the median expression of genes in sample 1.
b. plot a vertical line using abline().
17.3.2 Exercise 11b- bar plot + pie chart
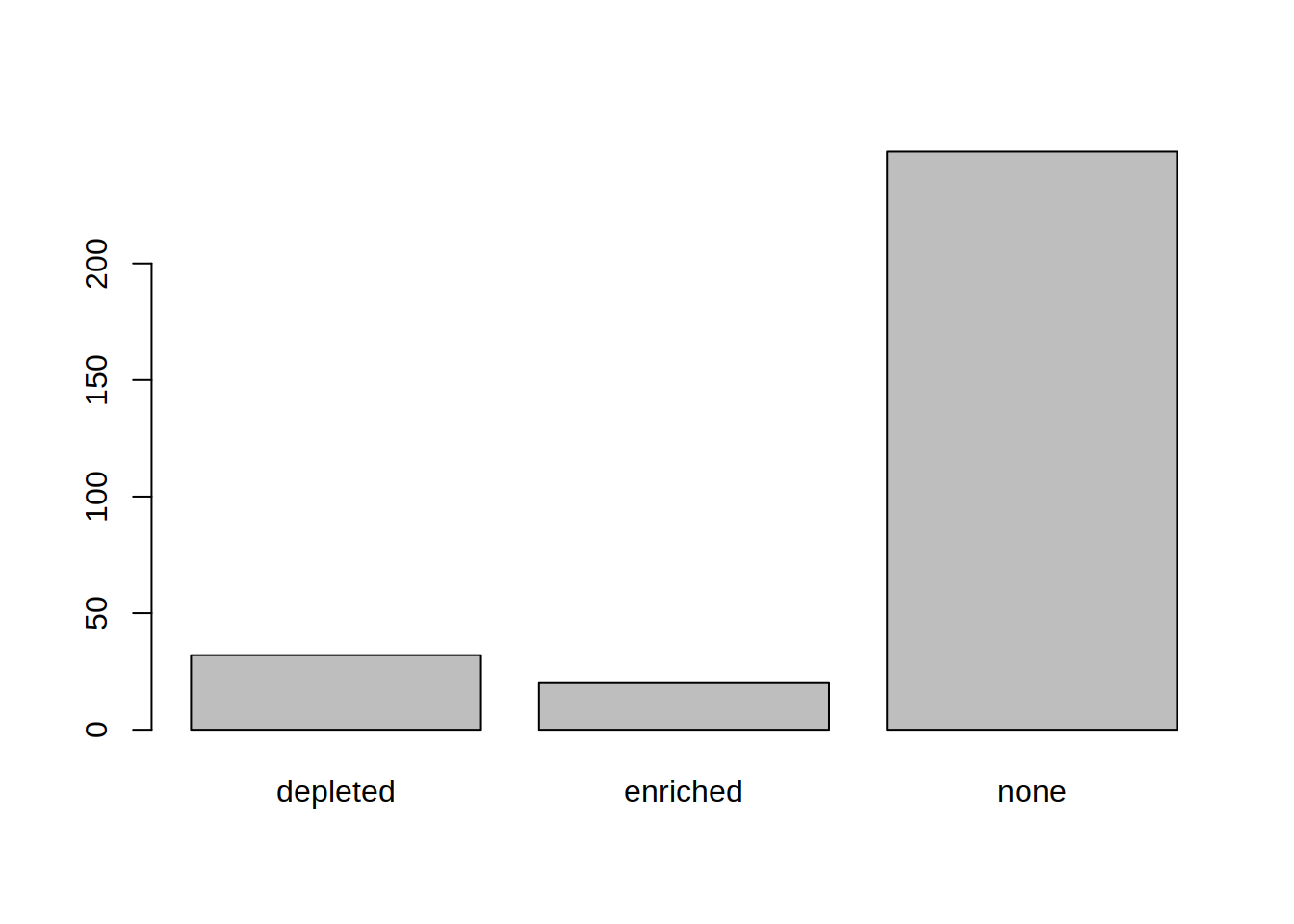
1- Create the following vector
2- The vector describes whether a gene is up- (enriched) or down- (depleted) regulated, or not regulated (none).
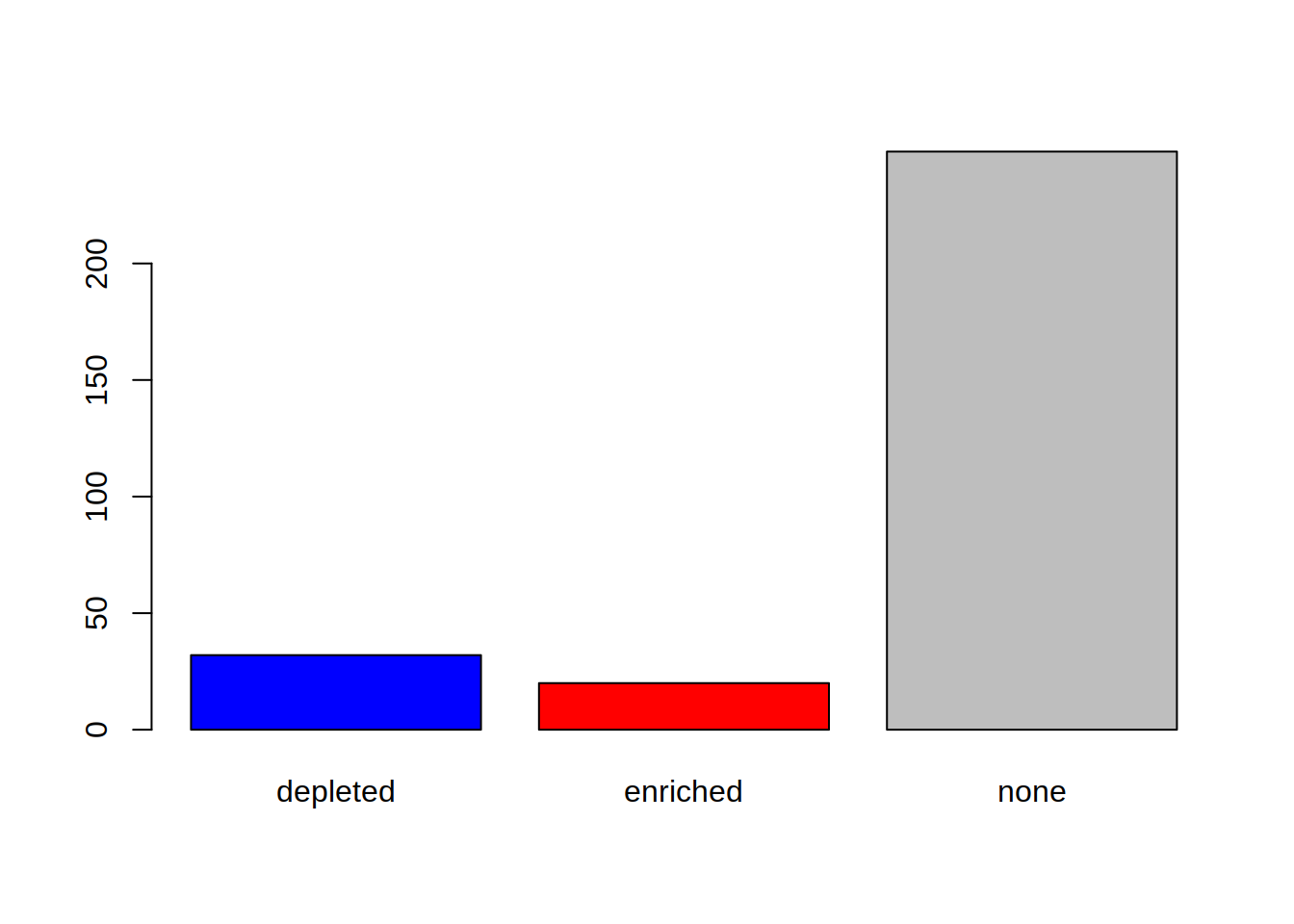
Produce a barplot that displays this information: how many genes are enriched, depleted, or not regulated.
3- Color the bars of the boxplot, each in a different color (3 colors of your choice)
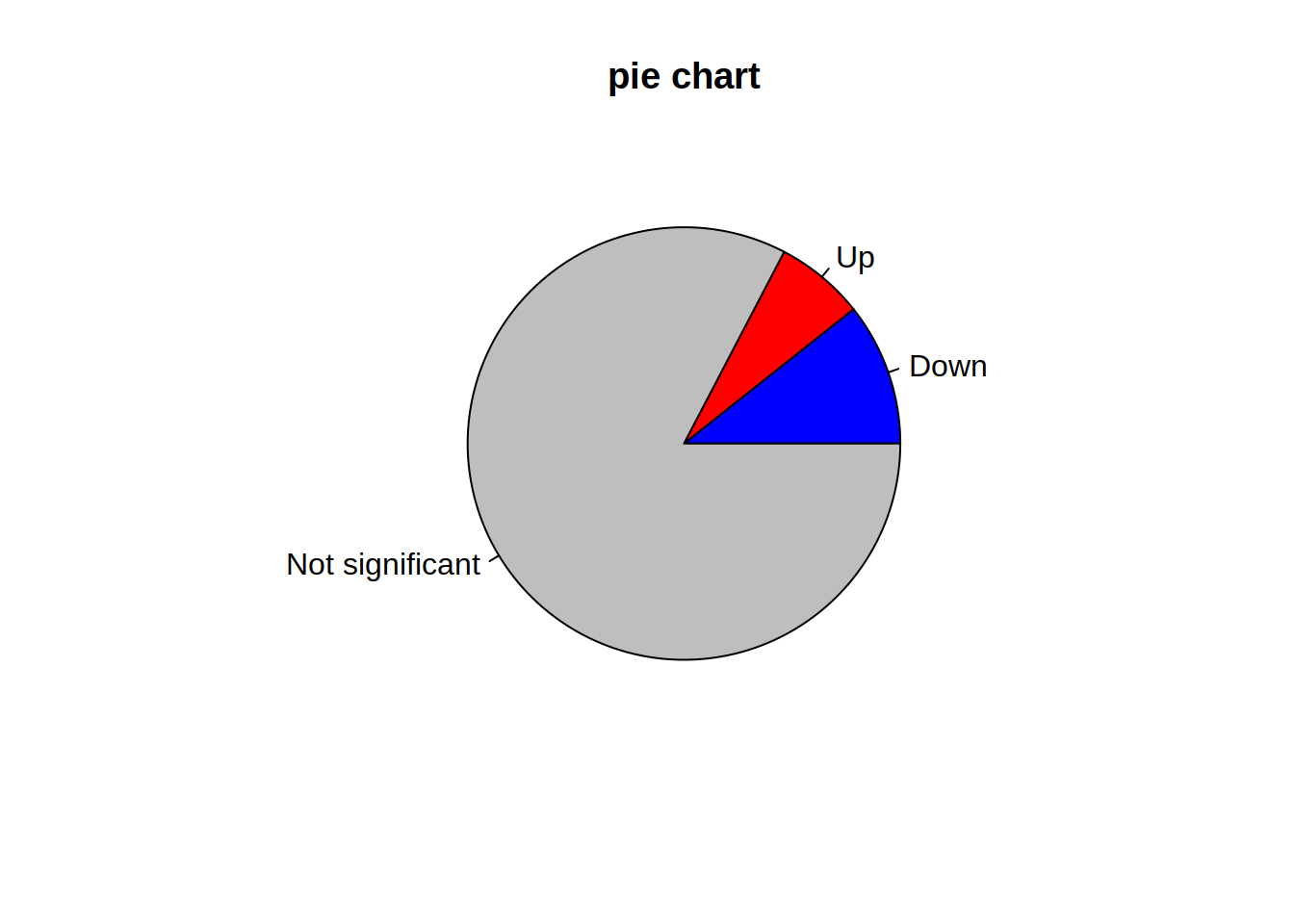
4- Use the argument “names.arg” in barplot() to rename the bars: Change depleted to “Down”, enriched to “Up”, none to “Not significant”
correction
barplot(table(genes_significance),
col=c("blue", "red", "grey"),
names.arg=c("Down", "Up", "Not significant"))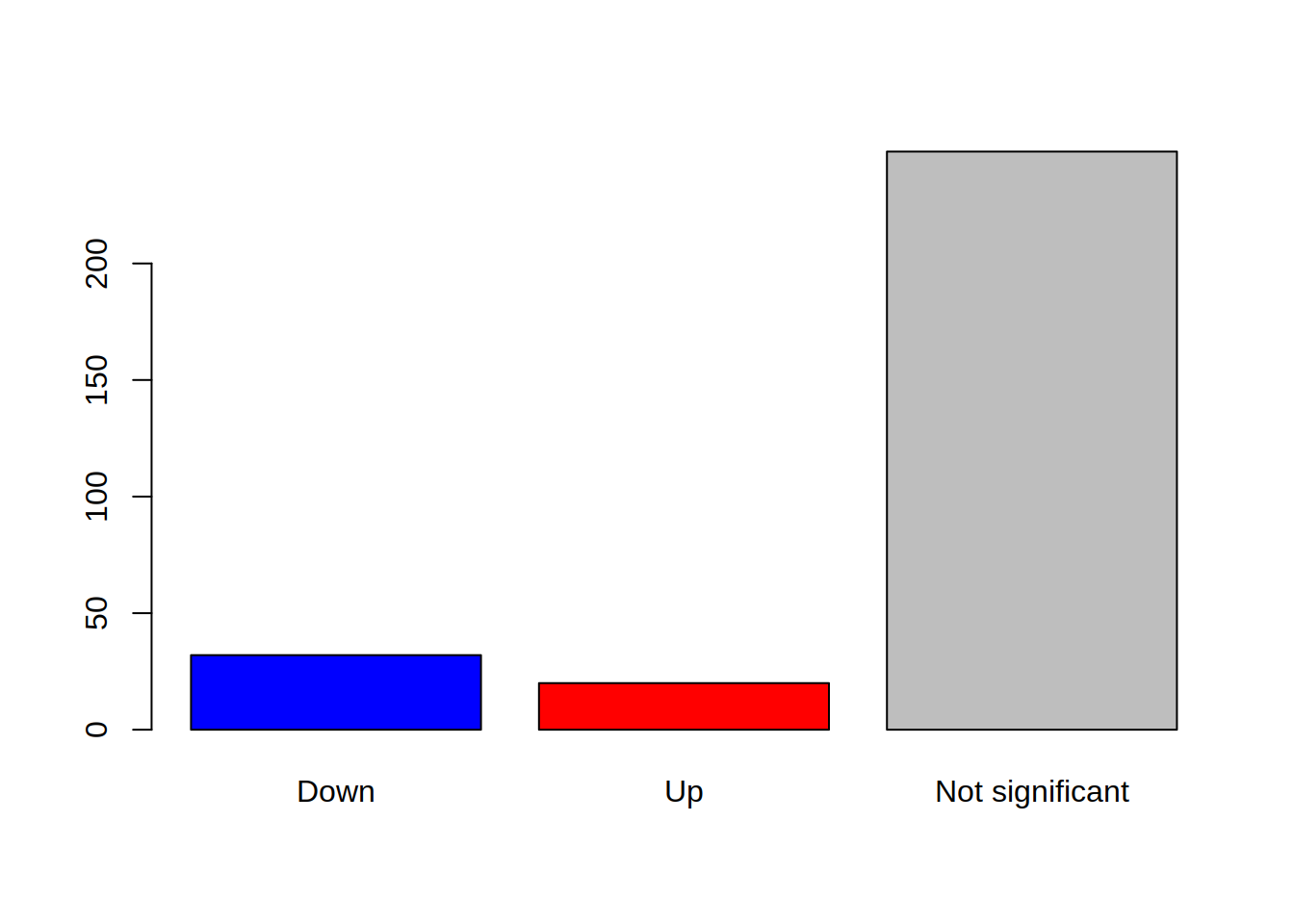
5- The “las” argument allow to rotate the x-axis labels for a better visibility. Try value 2 for las: what happens?
correction
barplot(table(genes_significance),
col=c("blue", "red", "grey"),
names.arg=c("Down", "Up", "Not significant"),
las=2)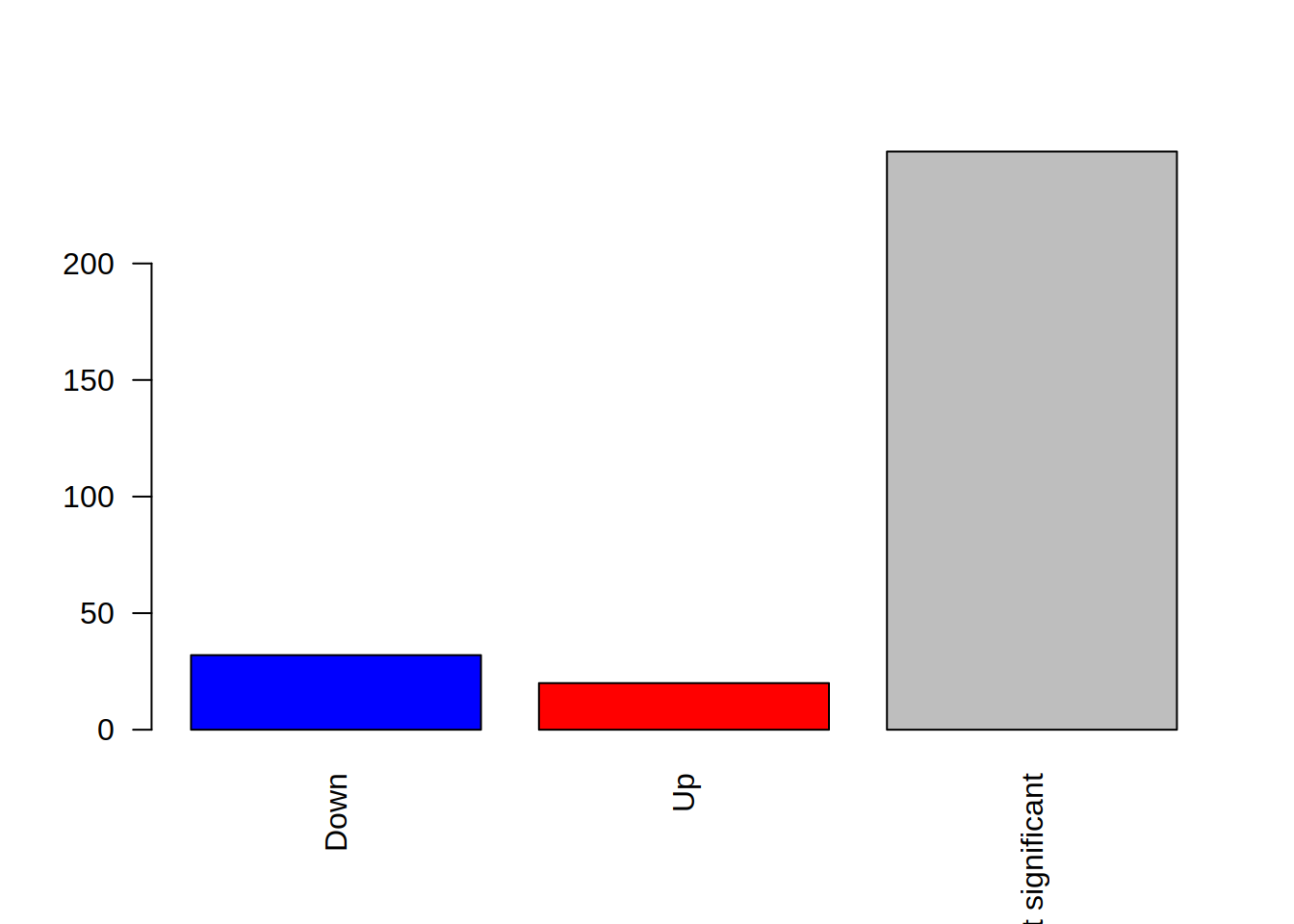
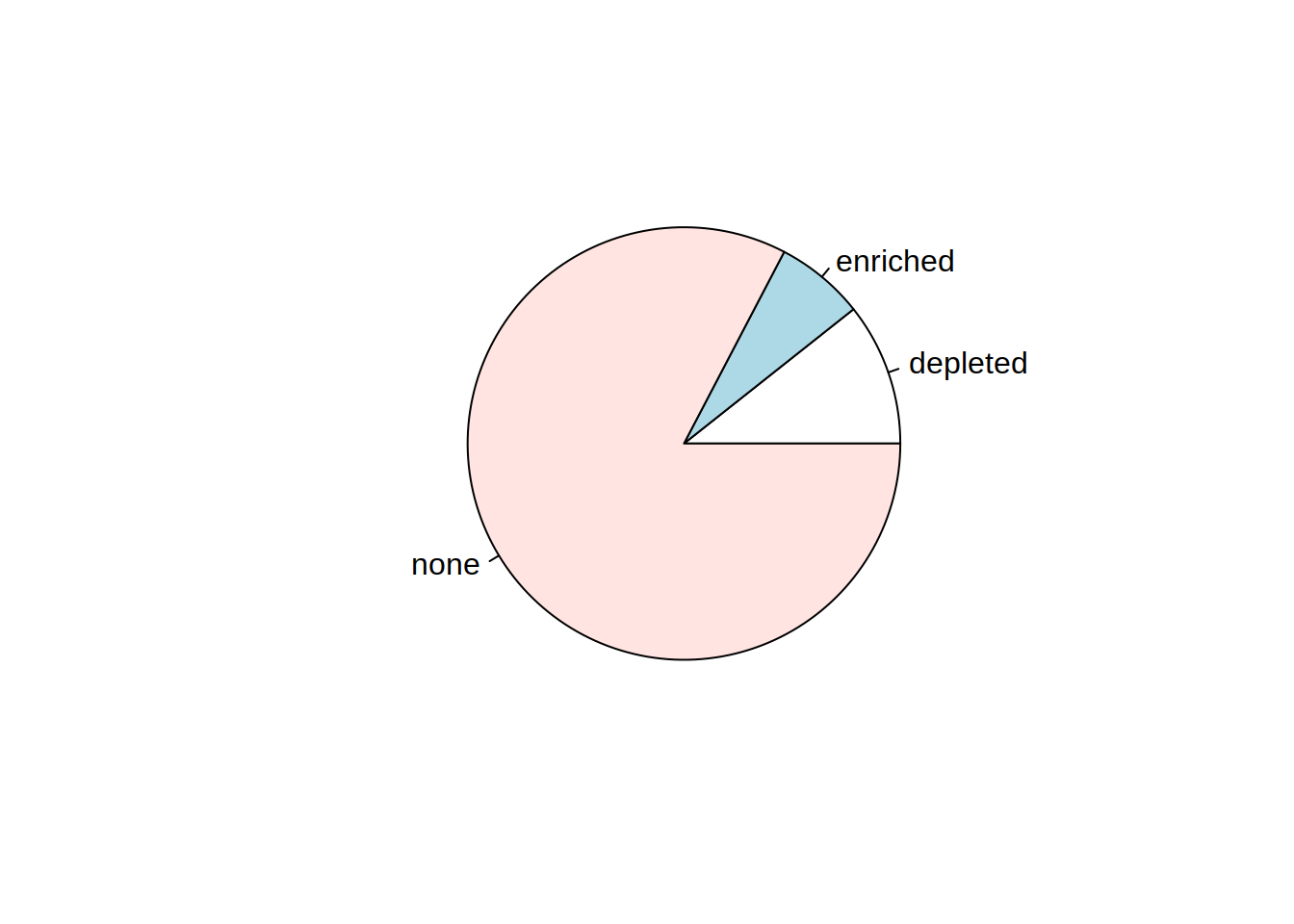
6- Create a pie chart of the same information (Enriched, Depleted, None)
Change the color of the slices, modify the labels, and add a title.
17.3.3 Exercise 11c- histogram
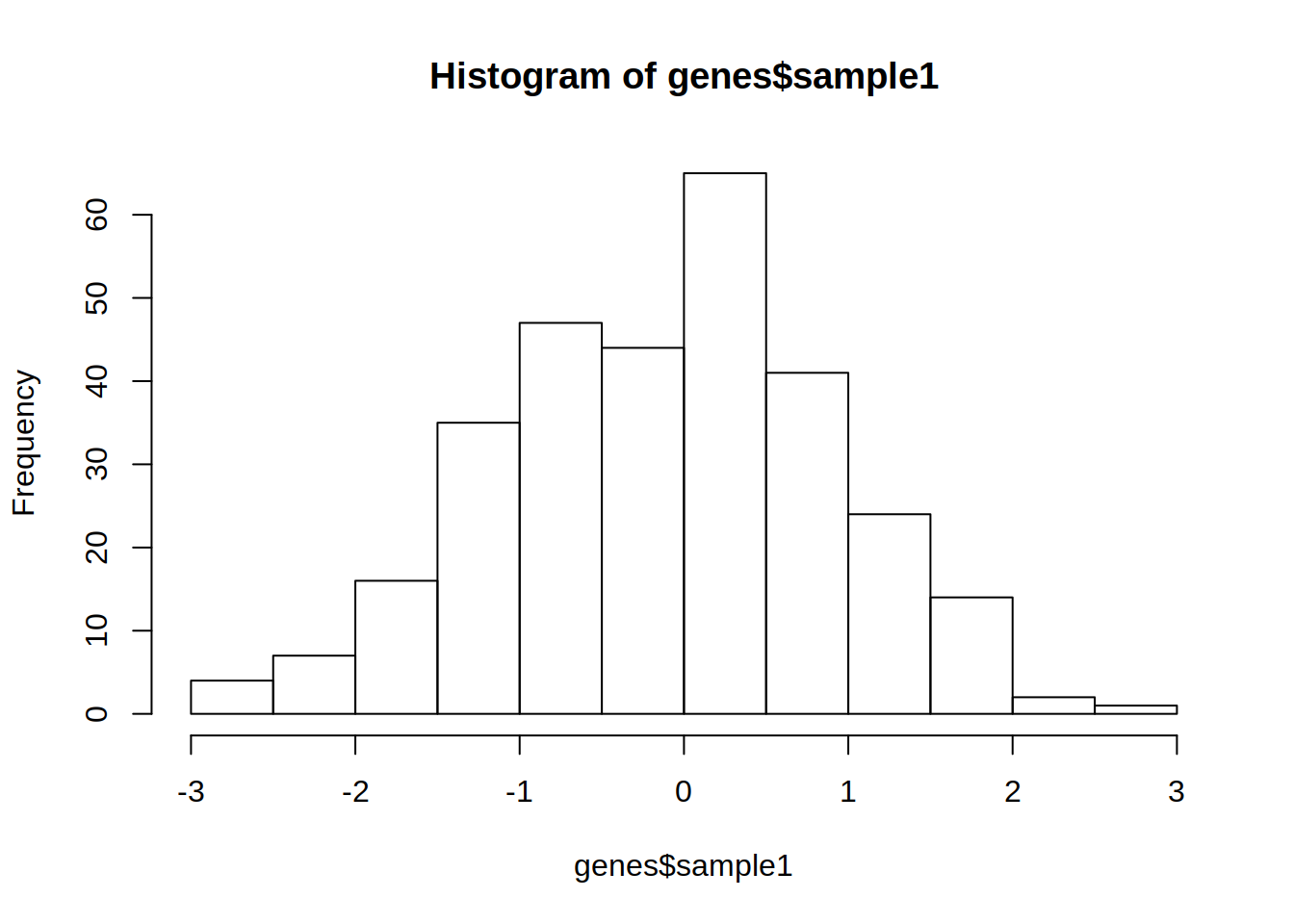
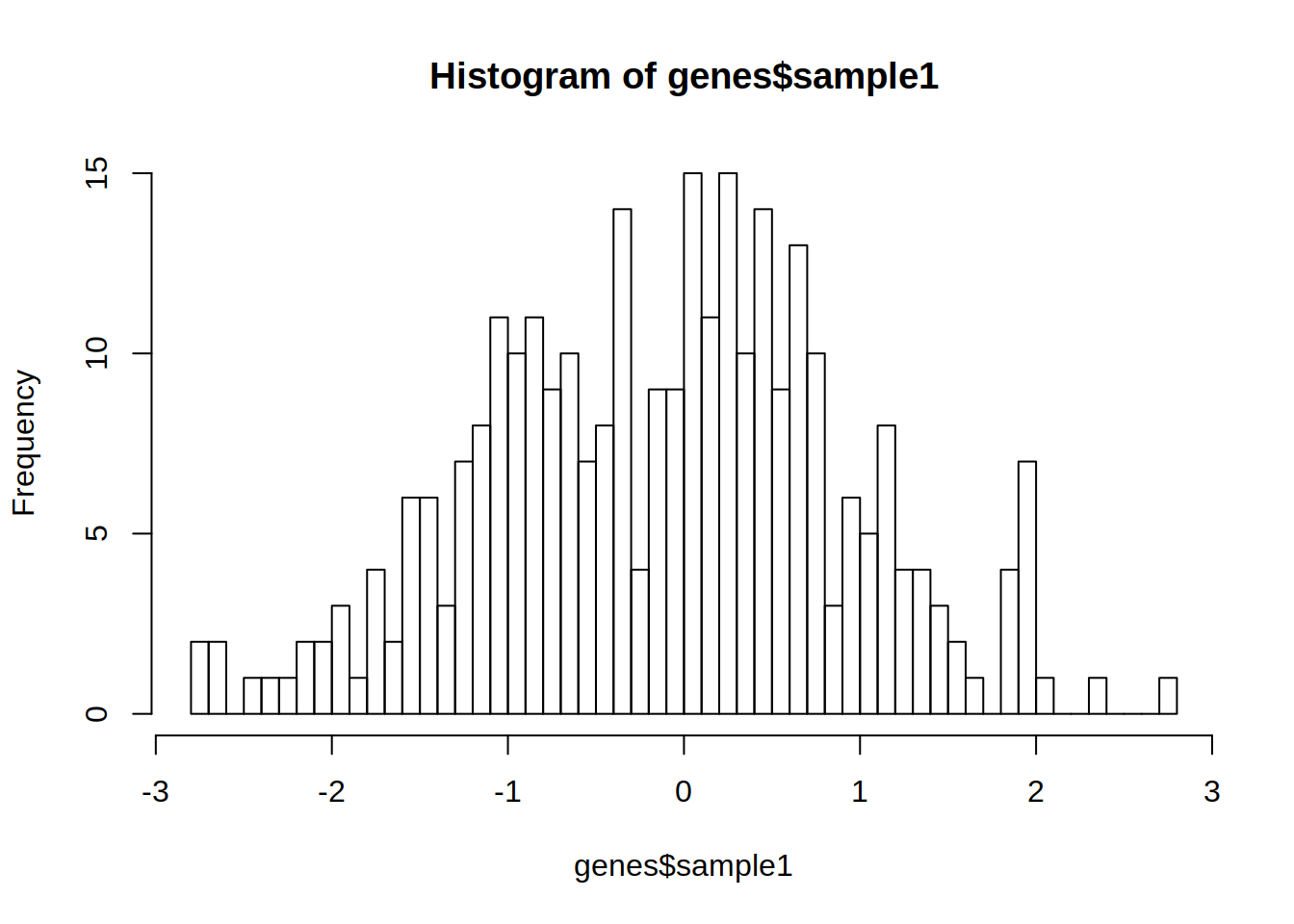
1- Use genes object from exercise 11a to create a histogram of the gene expression distribution of sample 1.
2- Repeat the histogram but change argument breaks to 50.
What is the difference ?
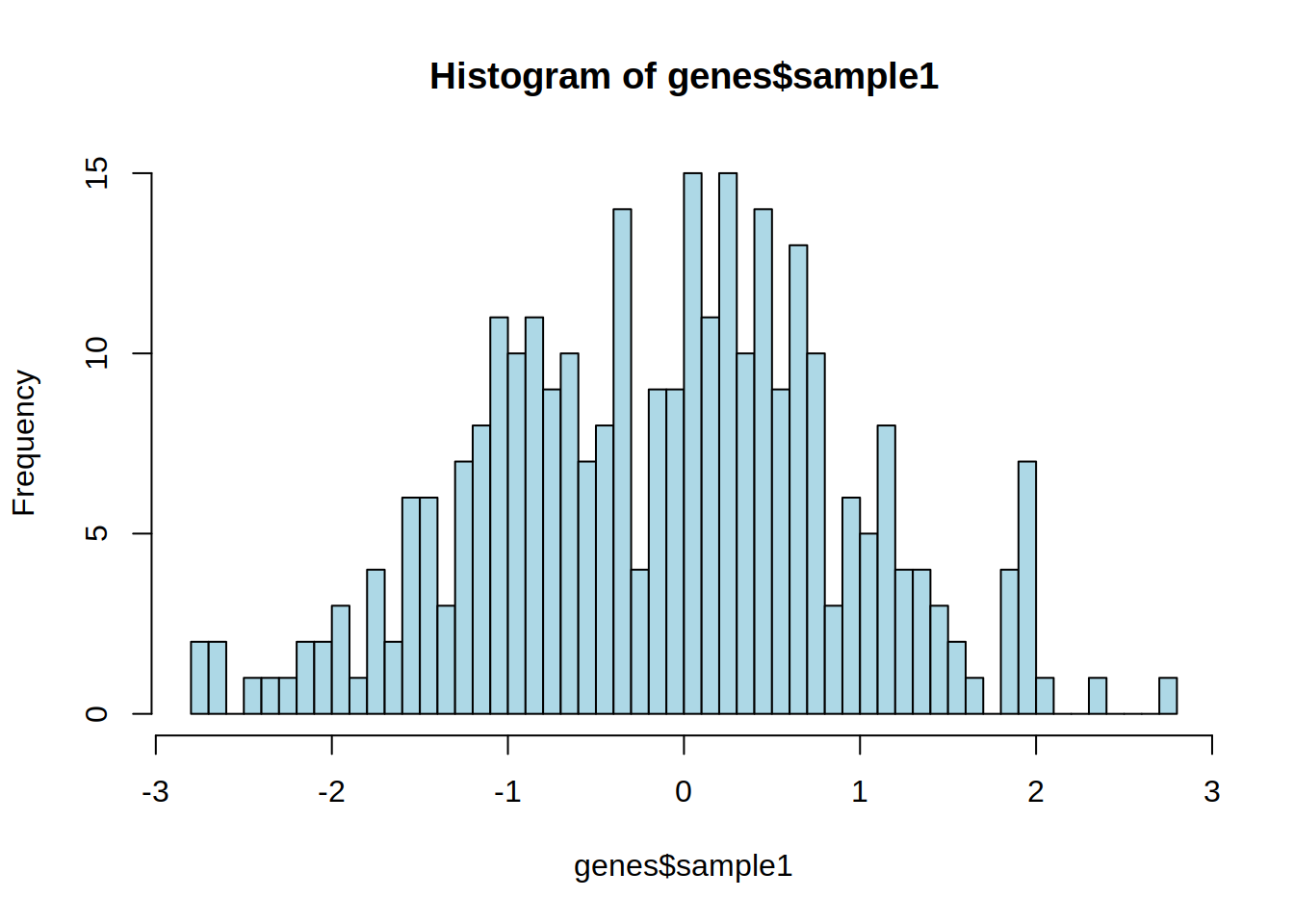
3- Color this histogram in light blue.
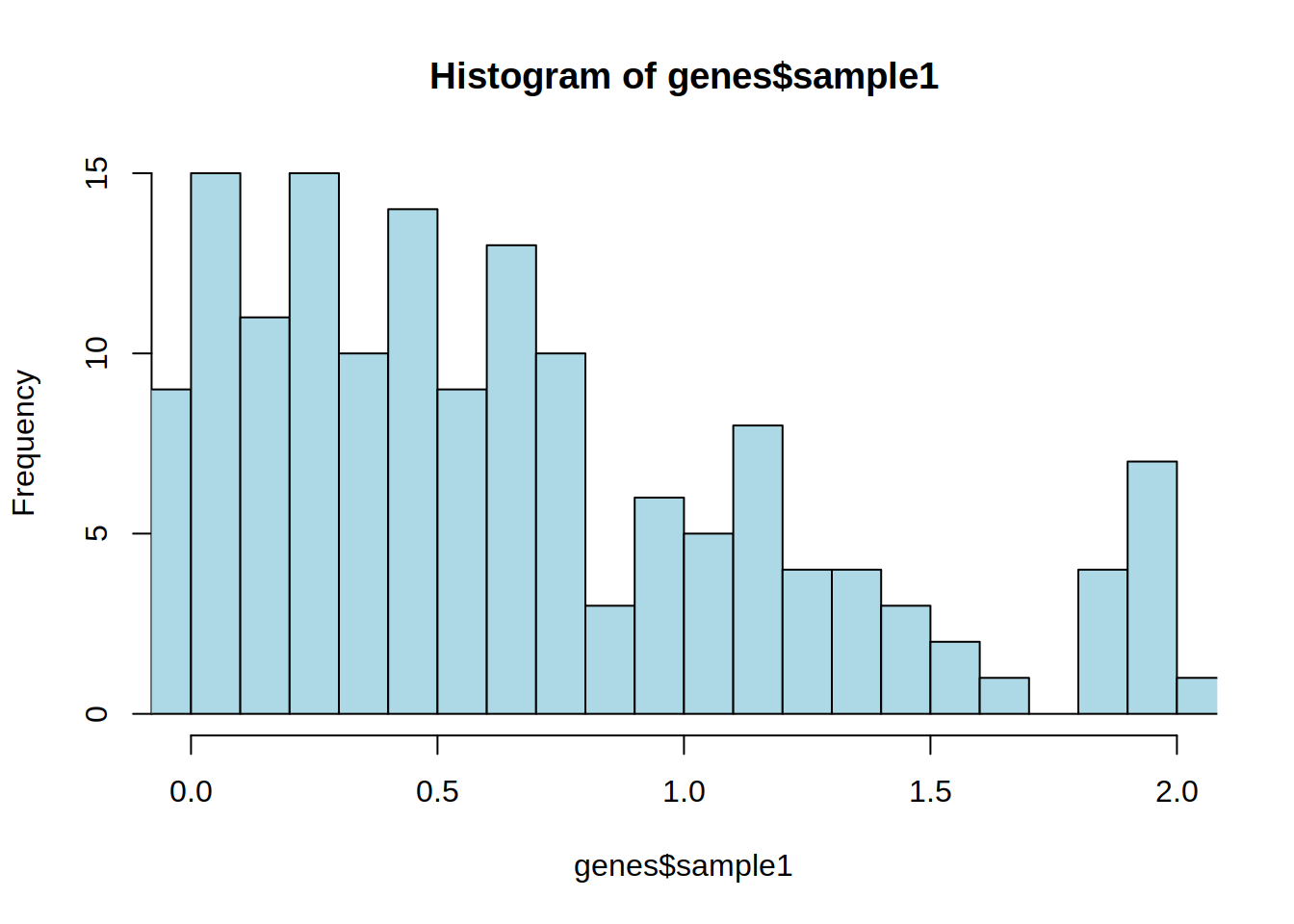
4- Zoom in the histogram: show only the distribution of expression values from 0 to 2 (x-axis) using the xlim argument.
5- Save the histogram in a pdf file.
correction
## png
## 2